5 plot with R
5.1 ggplot2
library(ggplot2)labs(x = NULL)can suppress x axis labelcalculated aesthetics via
stat()
ggplot(mpg, aes(displ)) +
geom_histogram(aes(y = stat(count)))
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.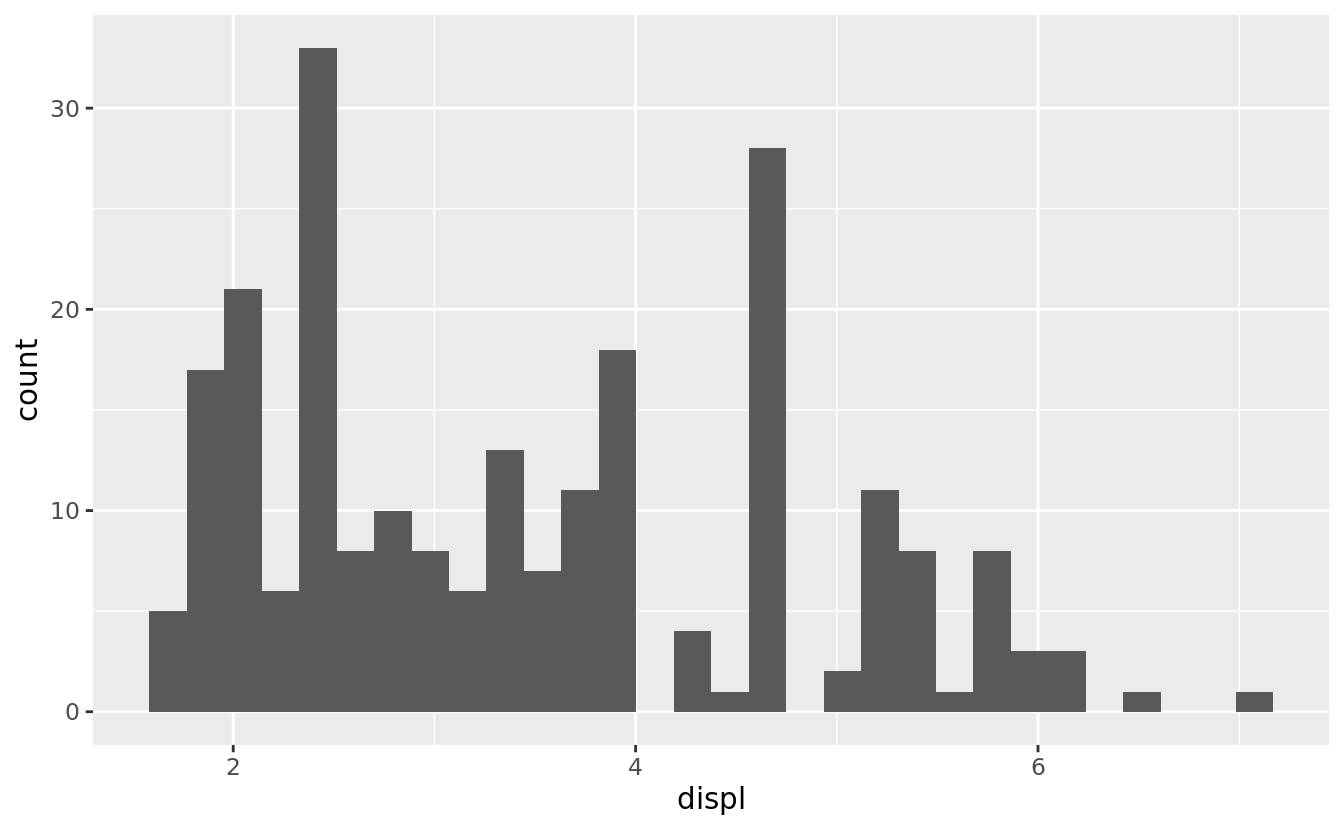
- color
scales::show_col(scales::hue_pal()(4))
- legend
ggplot(mtcars, aes(wt, mpg)) +
geom_point(aes(colour = factor(cyl), shape = factor(vs))) +
labs(x = "Weight (1000 lbs)", y = "Fuel economy (mpg)", colour = "Cylinders", shape = "Transmission") +
theme(legend.background = element_rect(colour = 'purple', fill = 'pink', size = 3, linetype='dashed'),
legend.margin = margin(20,15,10,5),
legend.box.background = element_rect(colour = 'blue', fill = 'yellow', size = 5, linetype='solid'),
legend.box.margin = margin(10,20,30,40))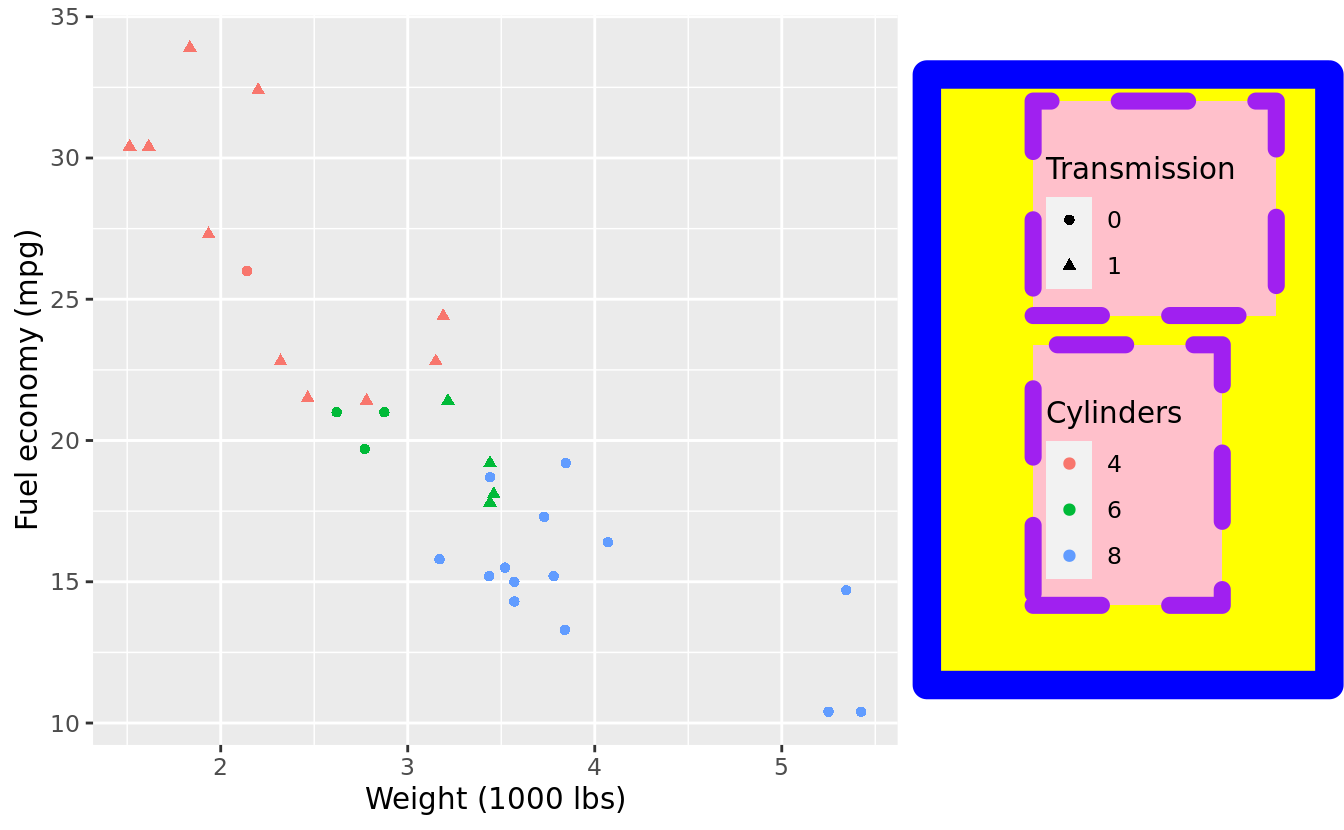
5.2 multi-plot
- 排列 plot,唯 patchwork 不破
we use cowplot::plot_grid() as reference
for gtable,
rbind()&cbind()just align plots, and you have to useggplotGrob()to convert ggplot object, examplefor egg,
ggarrange()doesn’t provide more feature, but some figure explains how to align panels: one & another 13for gridExtra,
grid.arrange()can’t add label, and syntax for layout is more complex. But can add title, contain hole, nest and multi-page vignettepatchwork provides a nice
+syntax, but it remains to see whether I have tolibrary()it, homemultipanelfigure ’s syntax is very different, but also very versatile, the author even use it to produce a supp figure of a biomedical paper guide
Thanks to this vignette, which contains many other useful tips:
- Plot insets
- share legend
- convert table to plot
following are R code I write when reproducing that vignette
gl <- lapply(1:4, function(ii)
grid::grobTree(grid::rectGrob(gp=grid::gpar(fill=ii, alpha=0.5)), grid::textGrob(paste0('plot', ii))))
grid.arrange(
grobs = gl,
widths = c(2, 1, 1),
layout_matrix = rbind(c(1, 2, NA),
c(3, 3, 4))
)
p1 <- qplot(mpg, wt, data = mtcars, colour = cyl)
p2 <- qplot(mpg, data = mtcars) + ggtitle("title")
p3 <- qplot(mpg, data = mtcars, geom = "dotplot")
p4 <-
p1 + facet_wrap( ~ carb, nrow = 1) + theme(legend.position = "none") +
ggtitle("facetted plot")
grid.arrange(
p1, p2, p3, p4,
widths = c(1, 1, 1),
layout_matrix = rbind(c(1, 2, 3),
c(4, 4, 4))
)
g <- ggplotGrob(qplot(1, 1) +
theme(plot.background = element_rect(colour = "black")))
qplot(1:10, 1:10) +
annotation_custom(
grob = g,
xmin = 1,
xmax = 5,
ymin = 5,
ymax = 10
) +
annotation_custom(
grob = grid::rectGrob(gp = grid::gpar(fill = "white")),
xmin = 7.5,
xmax = Inf,
ymin = -Inf,
ymax = 5
)5.3 graphics: prepare
# not avaliable in CRAN
if(!("nutshell.bbdb" %in% .packages(T))) remotes::install_github("cran/nutshell.bbdb")
if(!("nutshell.audioscrobbler" %in% .packages(T))) remotes::install_github("cran/nutshell.audioscrobbler")
if(!("nutshell" %in% .packages(T))) remotes::install_github("cran/nutshell")
if(!("learningr" %in% .packages(T))) remotes::install_github("cran/learningr")library(nutshell)
#> Loading required package: nutshell.bbdb
#> Loading required package: nutshell.audioscrobbler
library(learningr)
as_ggplot_list <- function(...) {
rlang::enexprs(...) %>% purrr::map(as.expression) %>% purrr::map(ggplotify::as.ggplot)
}
data(doctorates, batting.2008, turkey.price.ts, toxins.and.cancer, yosemite)
bat = plyr::mutate(
batting.2008,
PA = AB + BB + HBP + SF + SH,
OBP = (H + BB + HBP)/(AB + BB + HBP + SF)
)
bat.w.names <- plyr::mutate(
bat,
throws = as.factor(throws),
bats = as.factor(bats),
AVG = H/AB
)
toc = toxins.and.cancer
data(obama_vs_mccain, crab_tag)
obama_vs_mccain = obama_vs_mccain[!is.na(obama_vs_mccain$Turnout), ]
ovm = within(obama_vs_mccain, Region <- reorder(Region, Obama, median))5.4 graphics: univariate plot
5.4.1 pie chart
c(
"Fresh and forzen" = 7752,
"Reduced to meal, etc." = 1166,
"Canned" = 463,
"Cured" = 108
) %>% pie(main = "domestic catch 2006")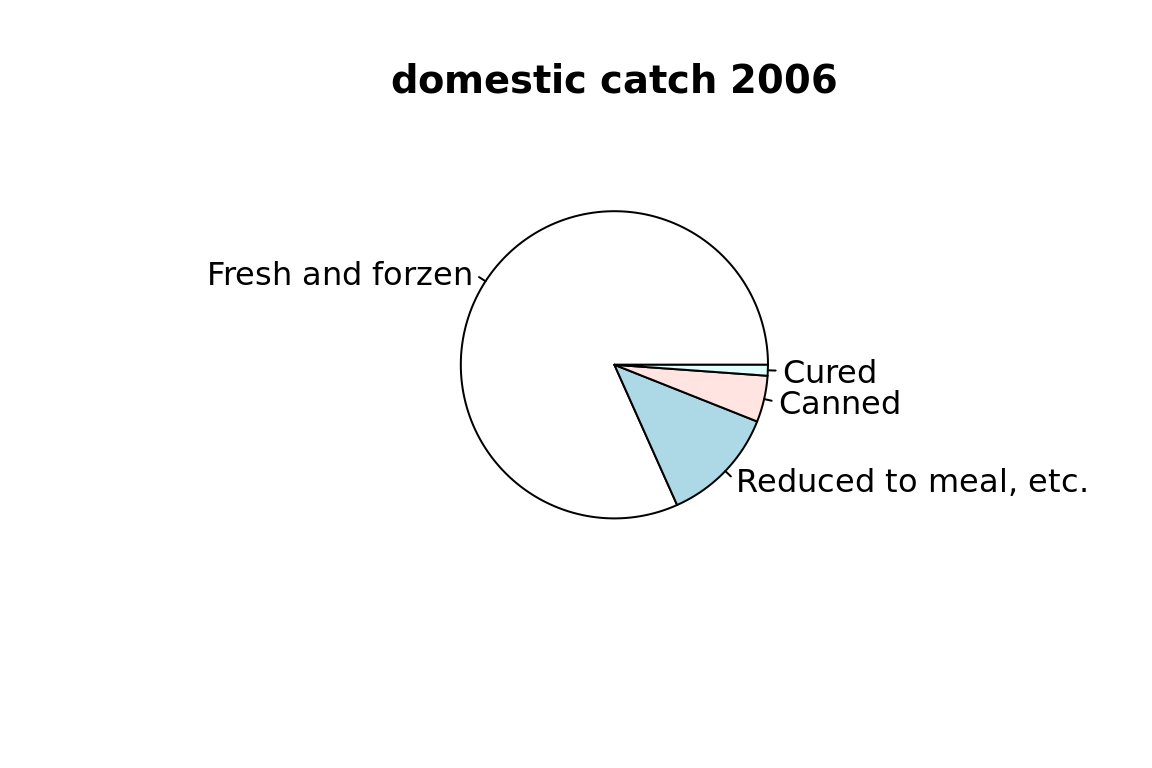
5.4.2 bar chart
transform data.frame into a matrix for barplot
doctorates.m <- as.matrix(doctorates[,-1])
rownames(doctorates.m) <- doctorates[,1]
doctorates.m
#> engineering science education health humanities other
#> 2001 5323 20643 6436 1591 5213 2159
#> 2002 5511 20017 6349 1541 5178 2141
#> 2003 5079 19529 6503 1654 5051 2209
#> 2004 5280 20001 6643 1633 5020 2180
#> 2005 5777 20498 6635 1720 5013 2480
#> 2006 6425 21564 6226 1785 4949 2436If the vector or matrix isn’t named, you need to use names.arg to specify the labels.
barplot(doctorates.m[1,])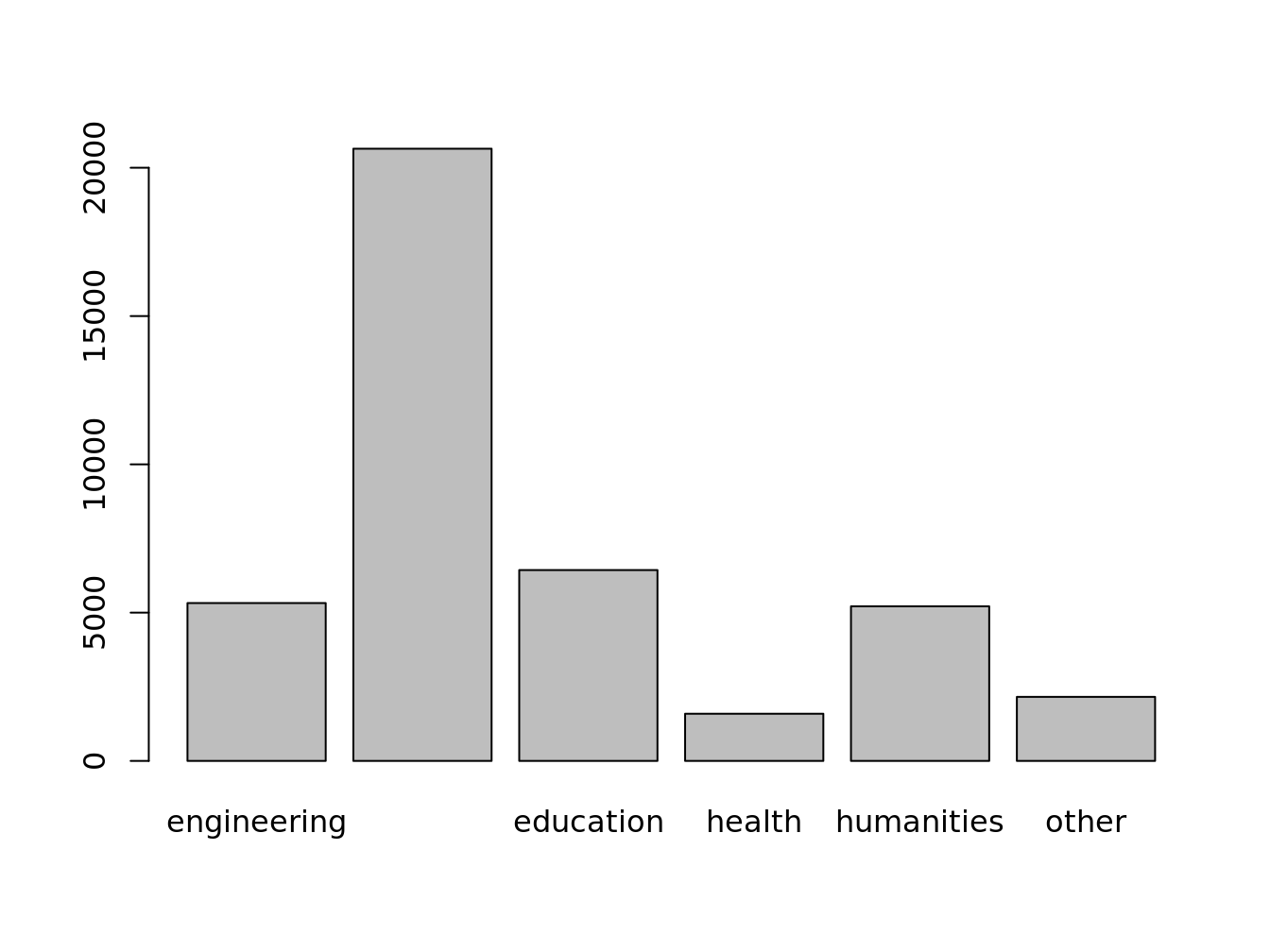
barplot(doctorates.m, beside = T, horiz = T, legend = T)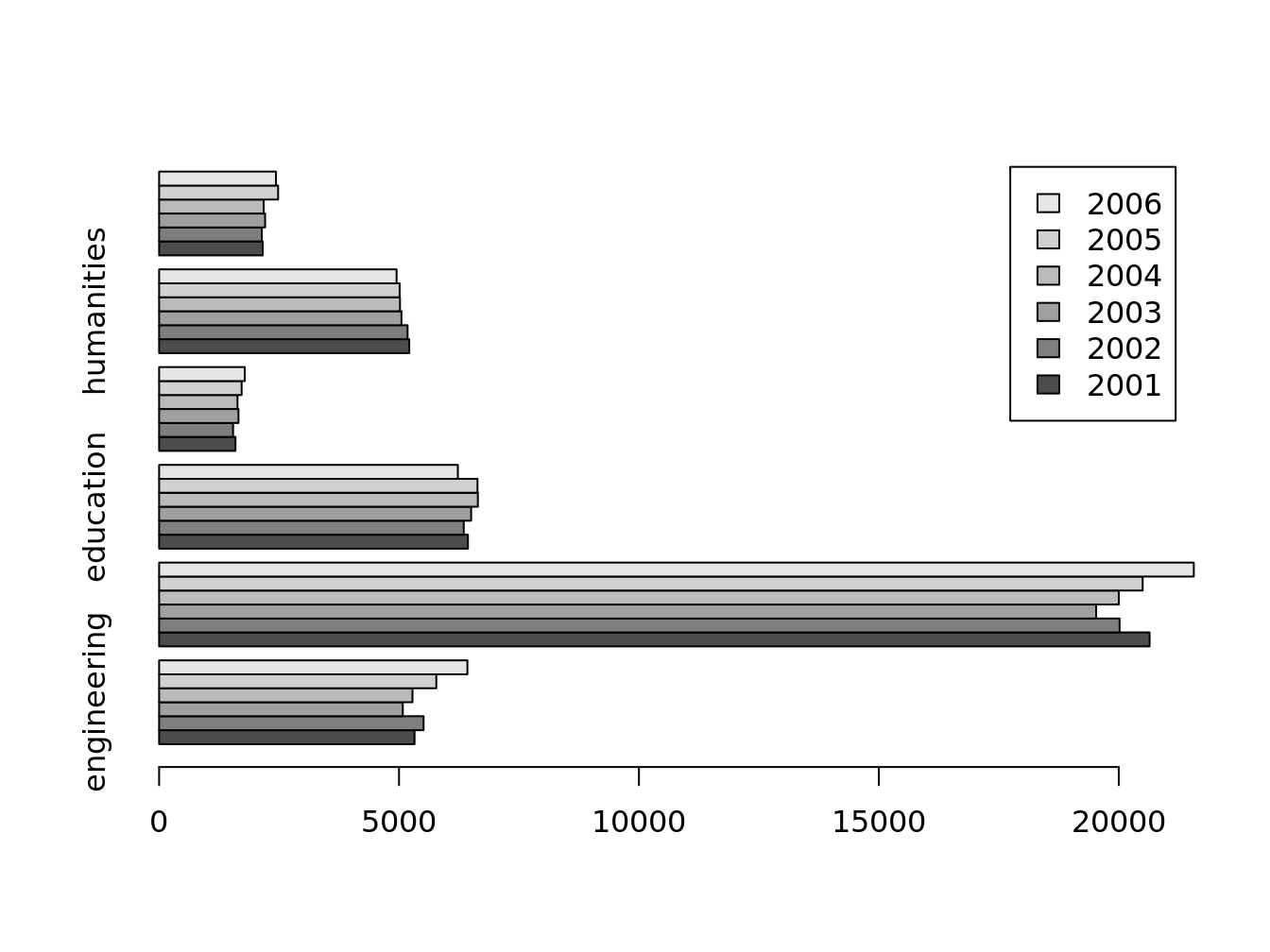
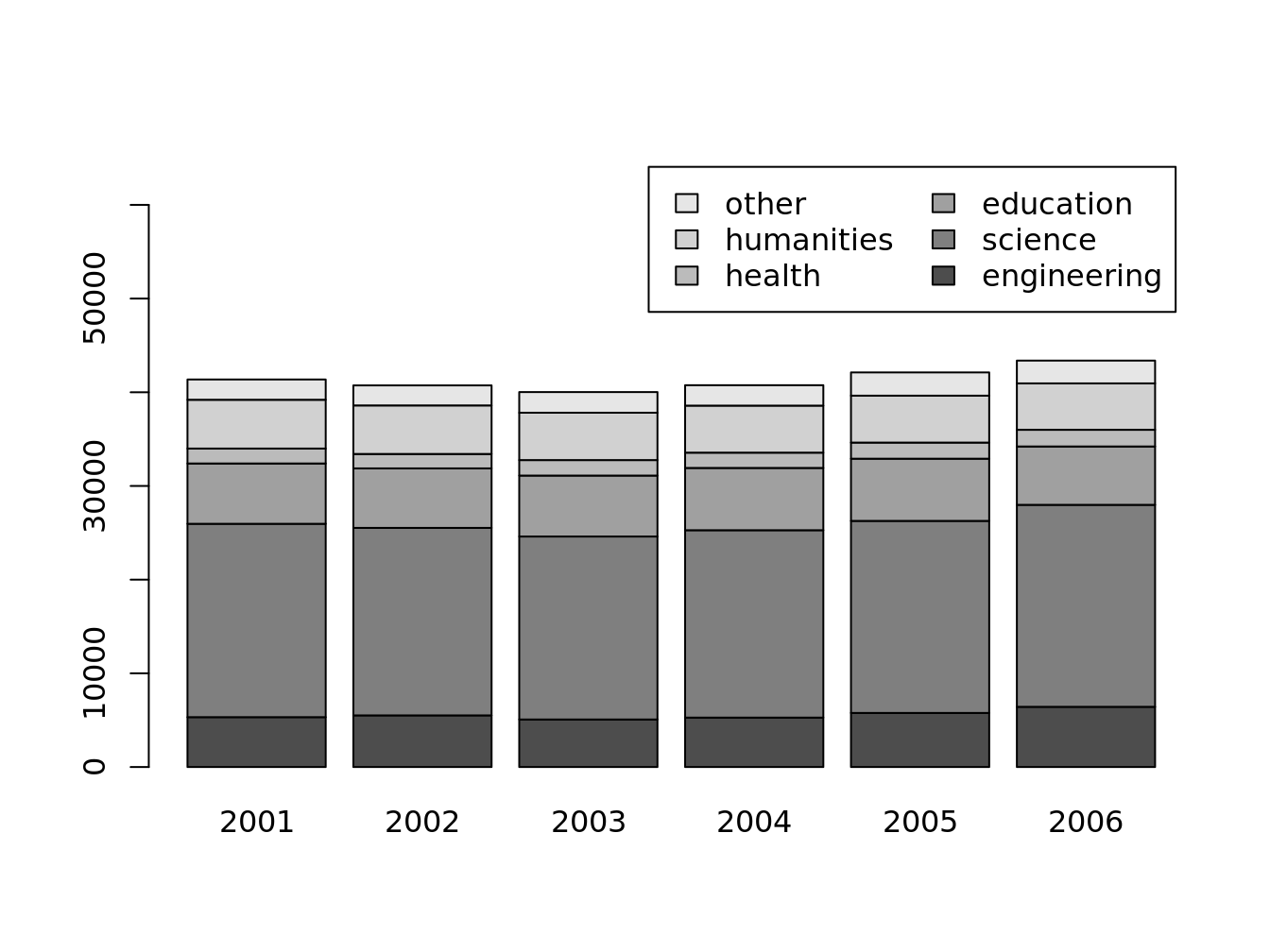
## we set `ylim` to make room for the legend
barplot(
t(doctorates.m), legend = T, ylim = c(0,66000),
args.legend = list(ncol = 2)
)
5.4.3 histogram
as_ggplot_list(
hist(obama_vs_mccain$Obama, main = 'number'),
hist(obama_vs_mccain$Obama, freq = F, main = 'frequency')
) %>% gridExtra::grid.arrange(grobs = ., nrow = 1)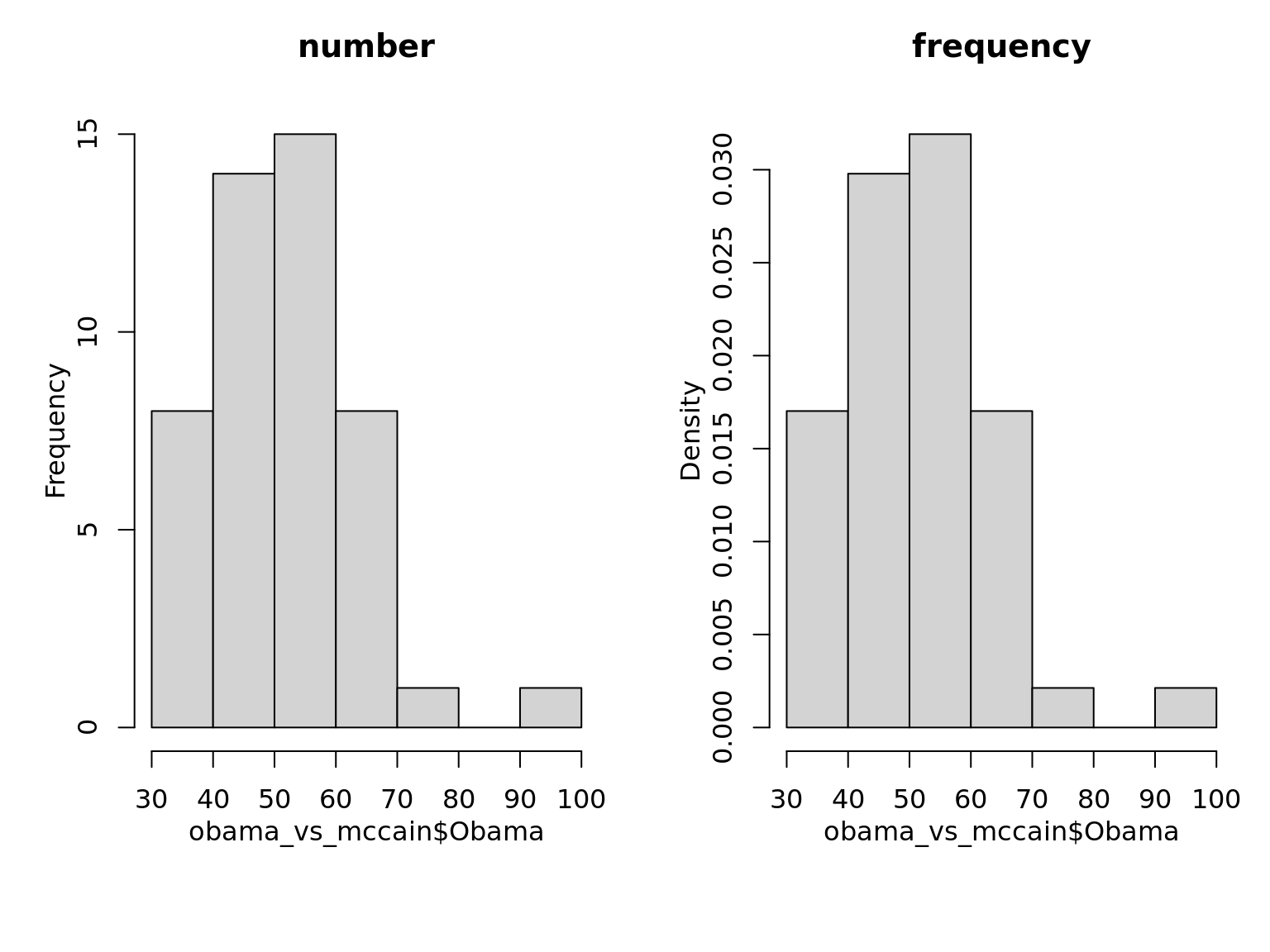
how to specify binwidth (breaks parameter)
hist(obama_vs_mccain$Obama, 4, main = "An exact number of bins")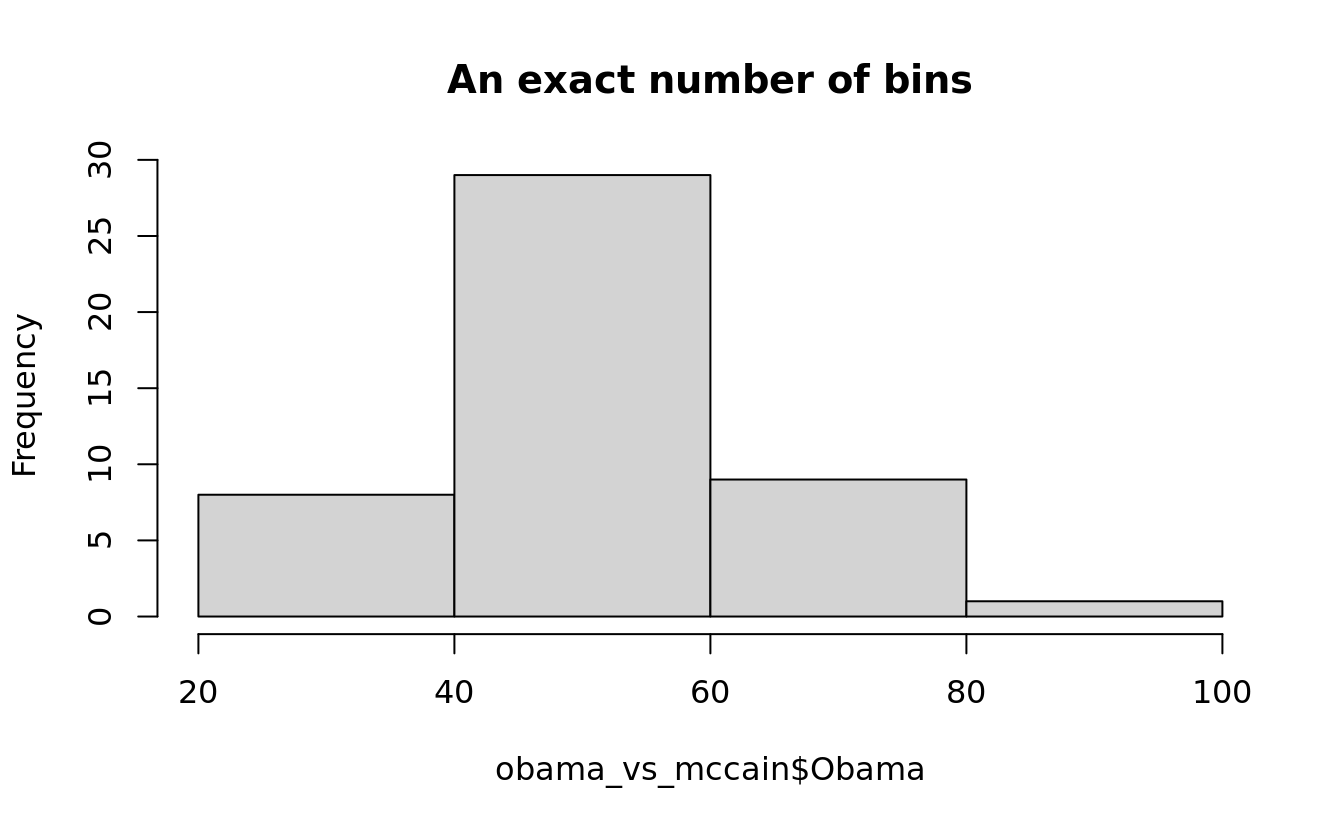
hist(obama_vs_mccain$Obama, seq.int(0, 100, 5), main = "A vector of bin edges")
hist(obama_vs_mccain$Obama, nclass.scott, main = "A function for the number of bins")
# you can also use the name of a method, such as `"FD"`
hist(obama_vs_mccain$Obama, function(x) {seq(min(x), max(x), length.out = 50)}, main = "A function for the bin edges")
5.5 graphics: bivariate plot
5.5.1 scatter plot
as_ggplot_list(
plot(obama_vs_mccain$Income, obama_vs_mccain$Turnout, col = "violet", pch = 20),
plot(obama_vs_mccain$Income, obama_vs_mccain$Turnout, log = "xy")
) %>% gridExtra::grid.arrange(grobs = ., nrow = 1)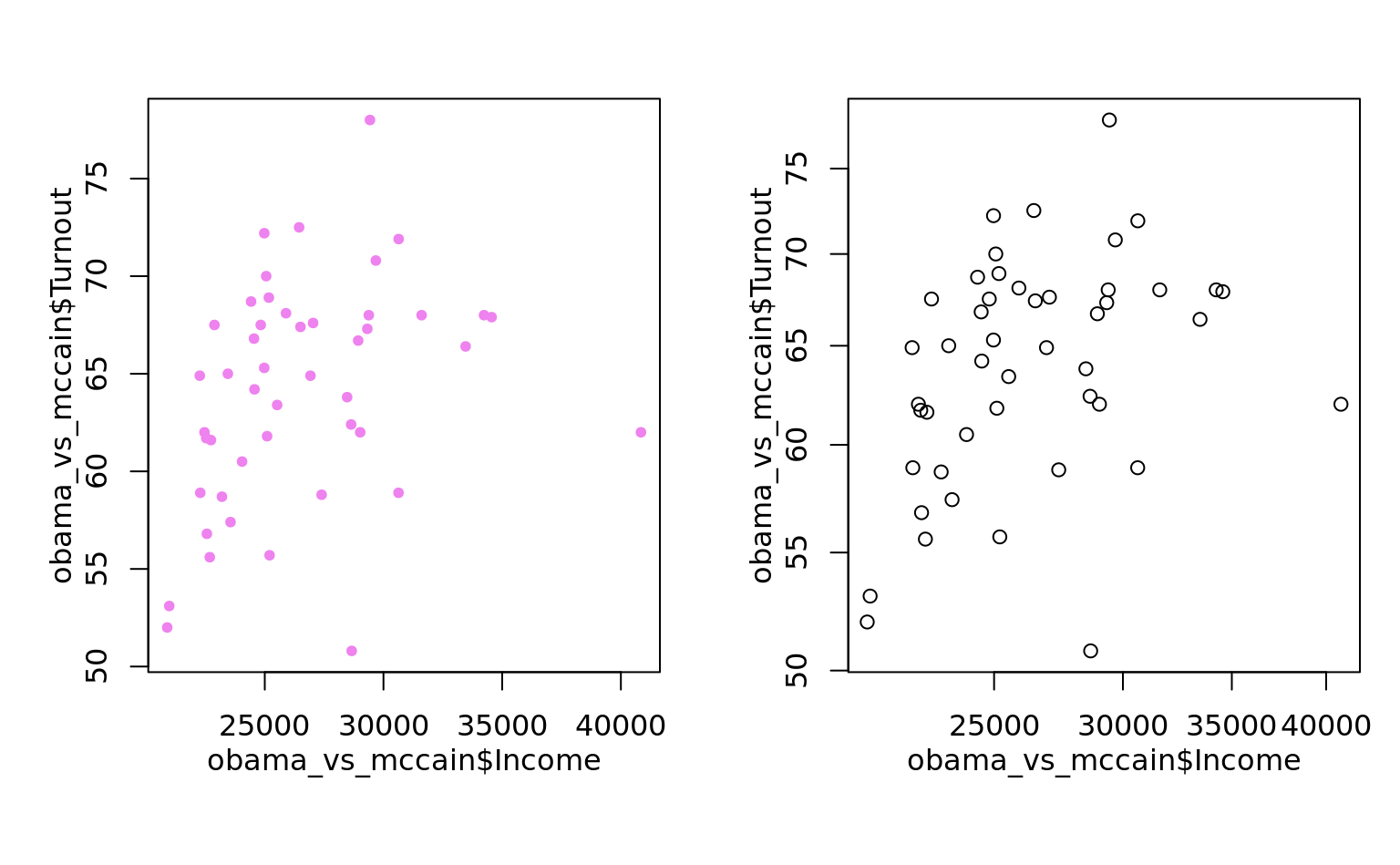
pairs of correlation
plot(bat[,c("H","R","SO","BB","HR")], main = '')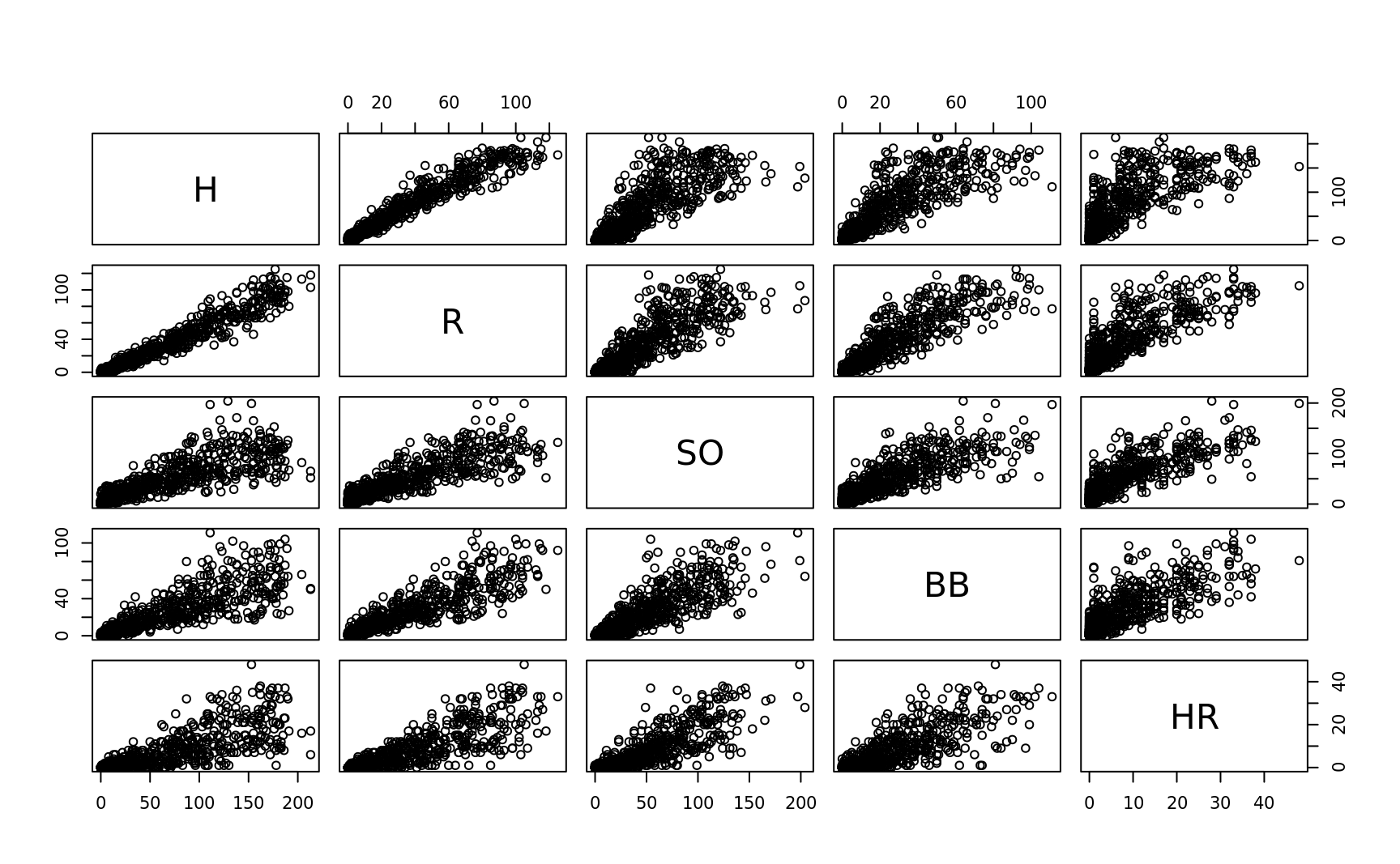
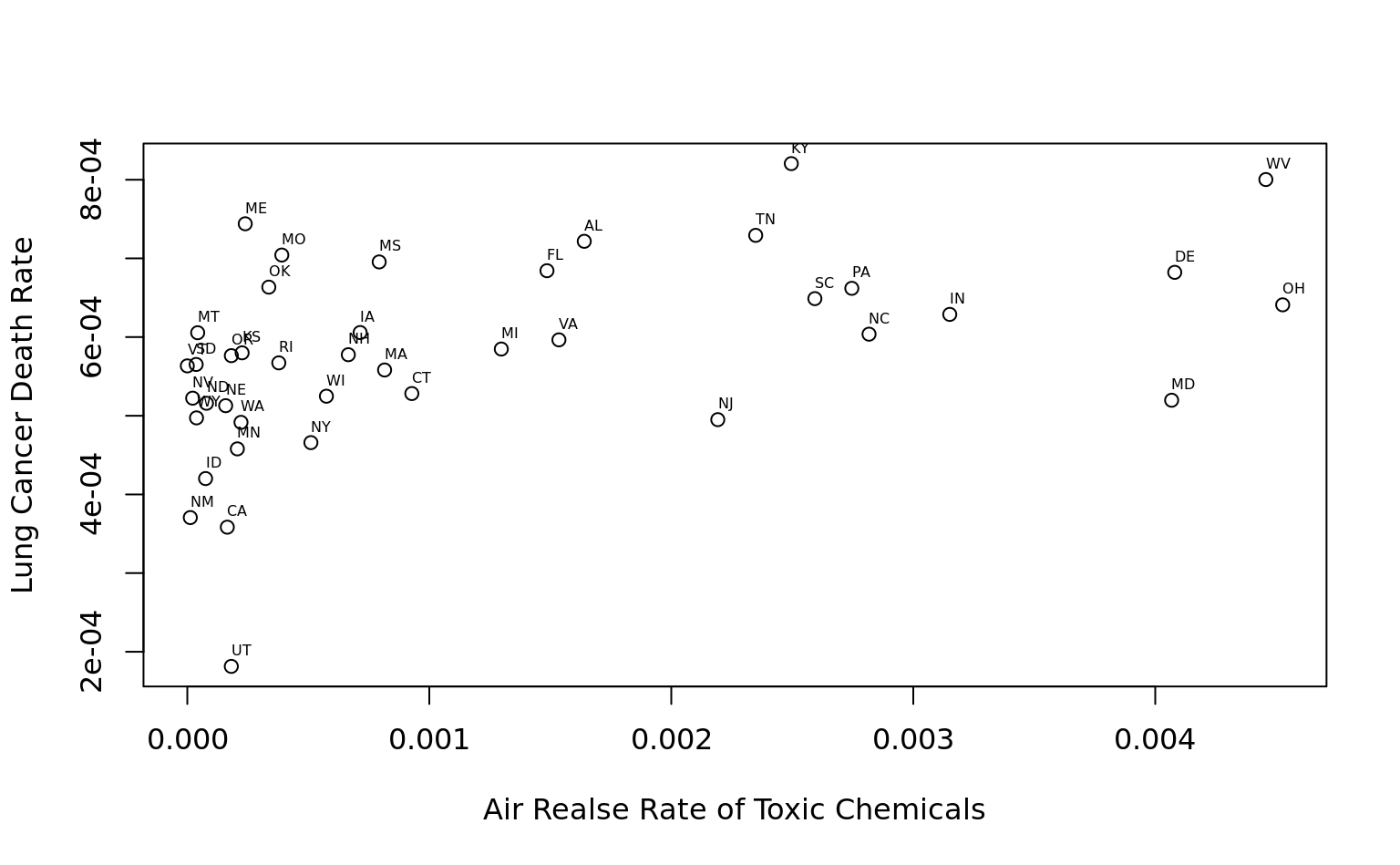
add text labels
plot(
toc$air_on_site/toc$Surface_Area, toc$deaths_lung/toc$Population,
xlab = "Air Realse Rate of Toxic Chemicals", ylab = "Lung Cancer Death Rate"
)
text(
toc$air_on_site/toc$Surface_Area, toc$deaths_lung/toc$Population,
cex = 0.5, labels = toc$State_Abbrev, adj = c(0, -1)
)
point shape is specified by pch (plotting character)
plot(1:25, pch = 1:25, bg = "blue")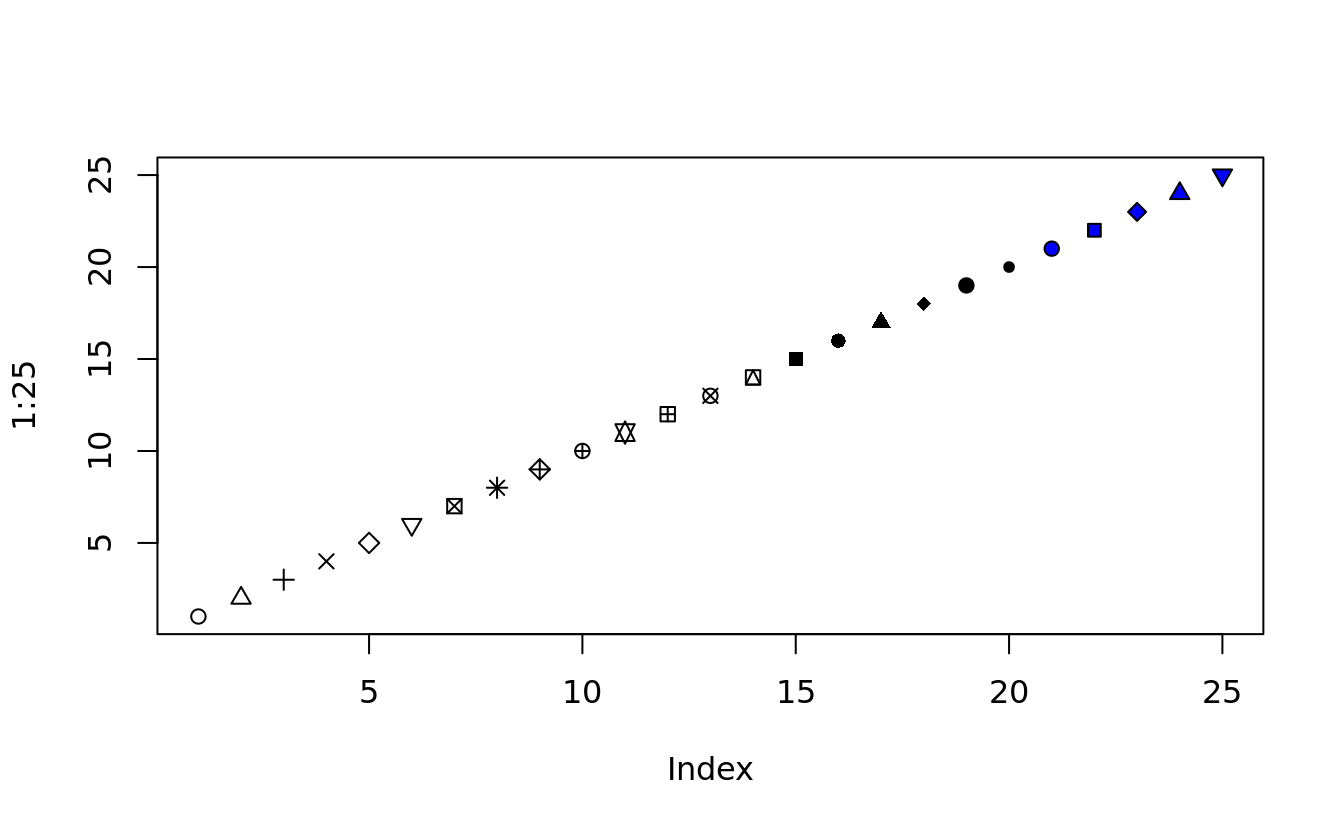
5.5.2 line plot
line plots are created in the same way as scatterplots, except that they take the argument type = “l”
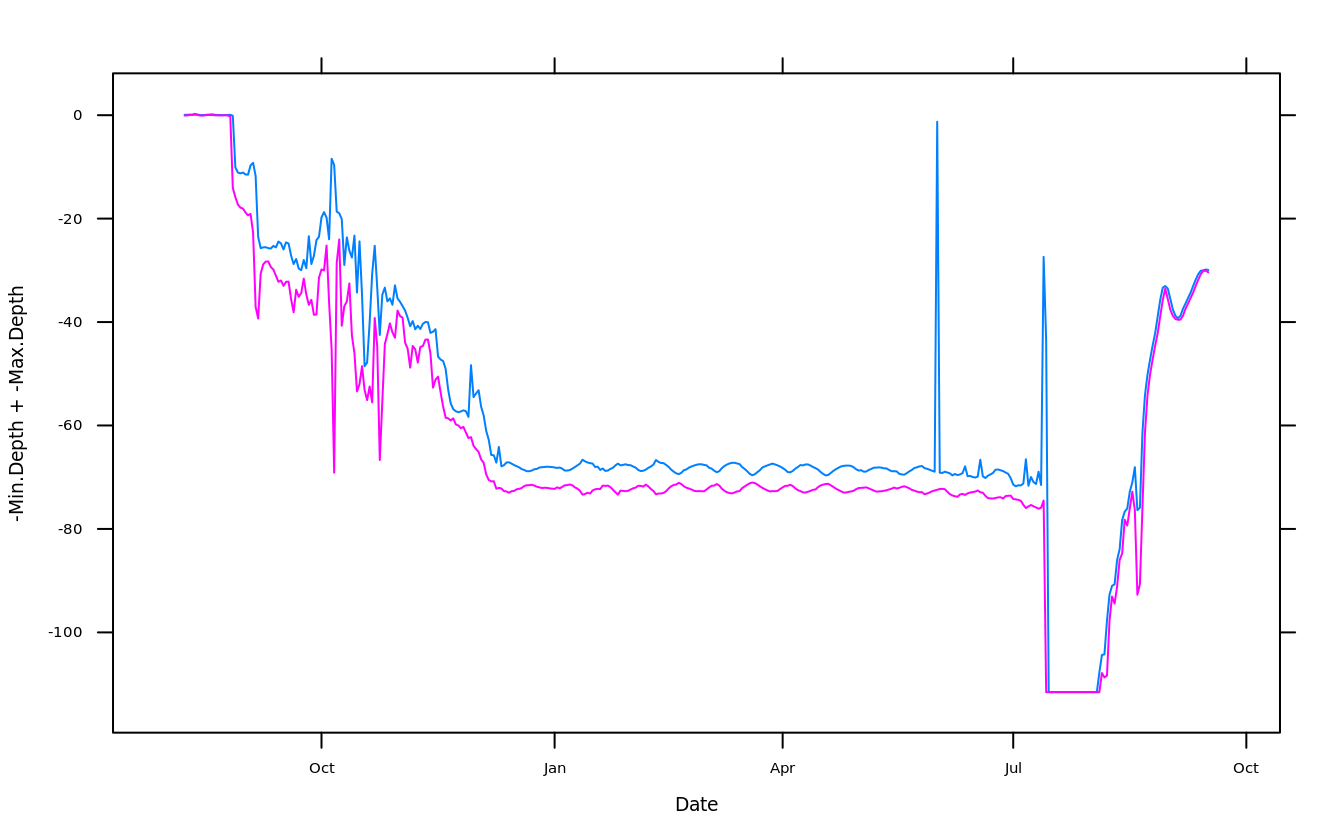
plot(crab_tag$daylog$Date, -crab_tag$daylog$Max.Depth, type = "l", ylim = c(-max(crab_tag$daylog$Max.Depth), 0))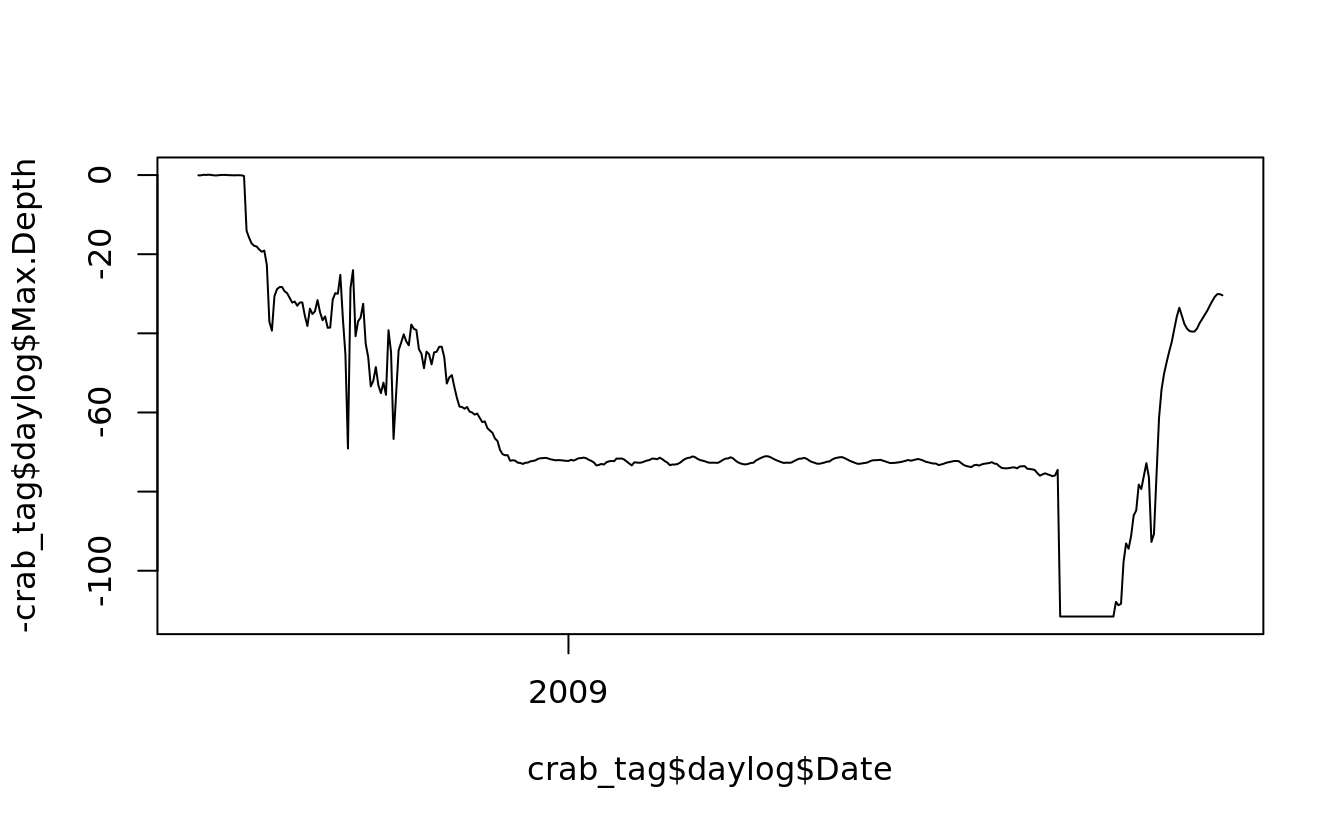
## use `lines()` to draw additional lines on an existing plot
lines(crab_tag$daylog$Date, -crab_tag$daylog$Min.Depth, col = "blue")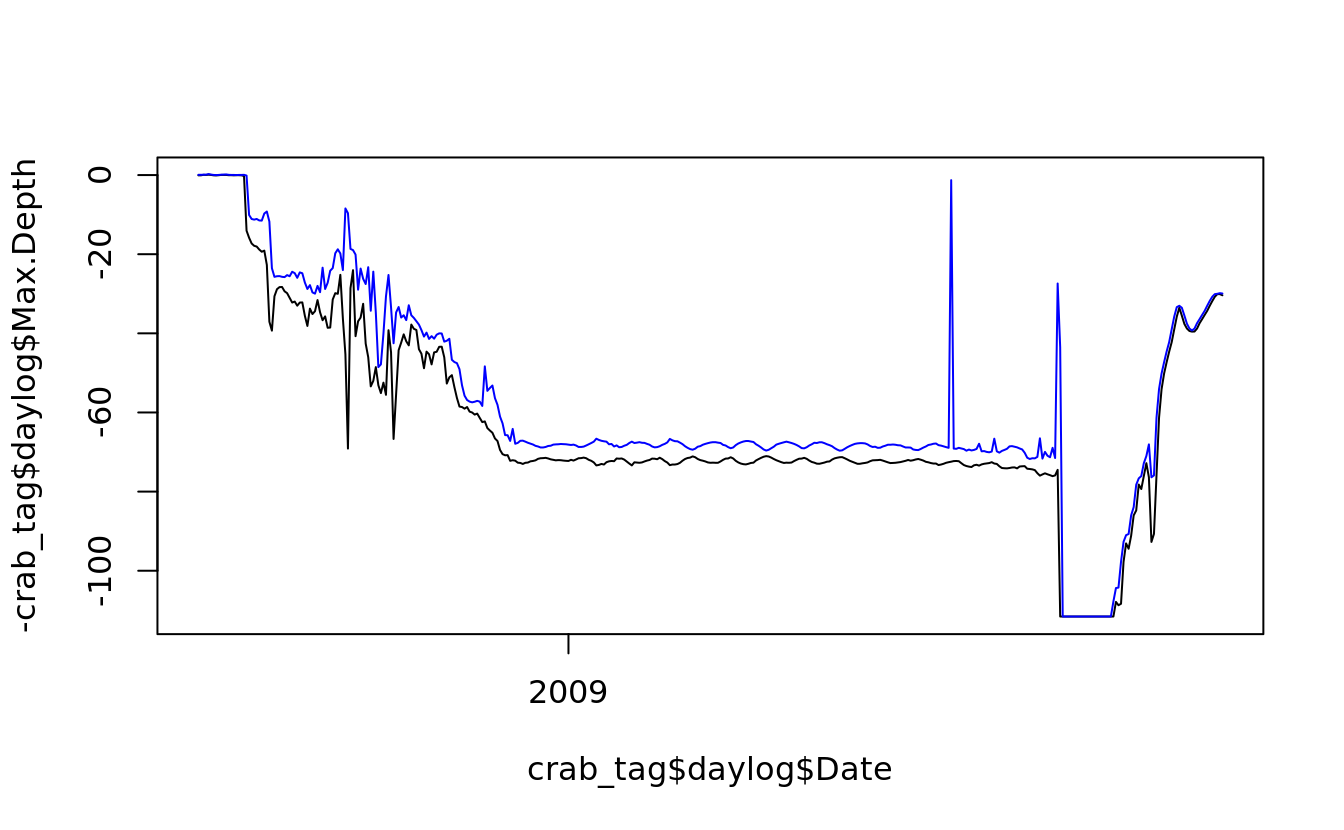
5.5.3 box plot
- The box shows the interquartile range, which contains values between the 25th and 75th percentile;
- The line inside the box shows the median;
- The two whiskers on either side of the box show the adjacent values, which are intended to show extreme values;
- When there are values far outside the range we would expect for normally distributed data, those outlying values are plotted separately.
- Specifically, here is how the adjacent values are calculated:
- the upper adjacent value is the value of the largest observation that is less than or equal to the upper quartile plus 1.5 times the length of the interquartile range;
- the lower adjacent value is the value of the smallest observation that is greater than or equal to the lower quartile less 1.5 times the length of the interquartile range.
as_ggplot_list(
boxplot(Obama ~ Region, data = obama_vs_mccain),
boxplot(Obama ~ Region, within(obama_vs_mccain, Region <- reorder(Region, Obama, median)))
) %>% gridExtra::grid.arrange(grobs = ., nrow = 1)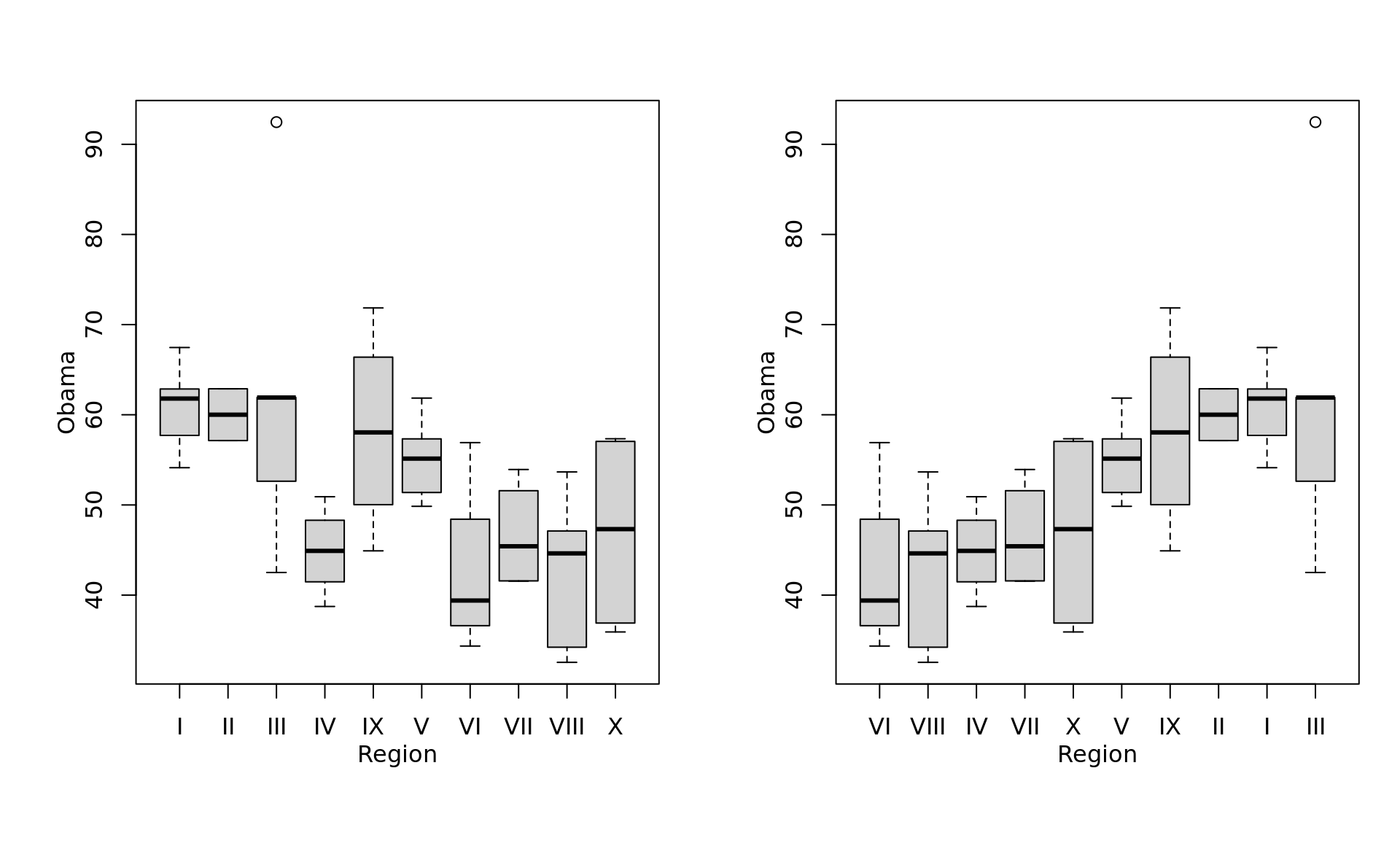
5.5.4 plot categorical data
# conditional density of a set of categories dependent on a numeric value
cdplot(bats ~ AVG, bat.w.names, subset = (bat.w.names$AB > 100))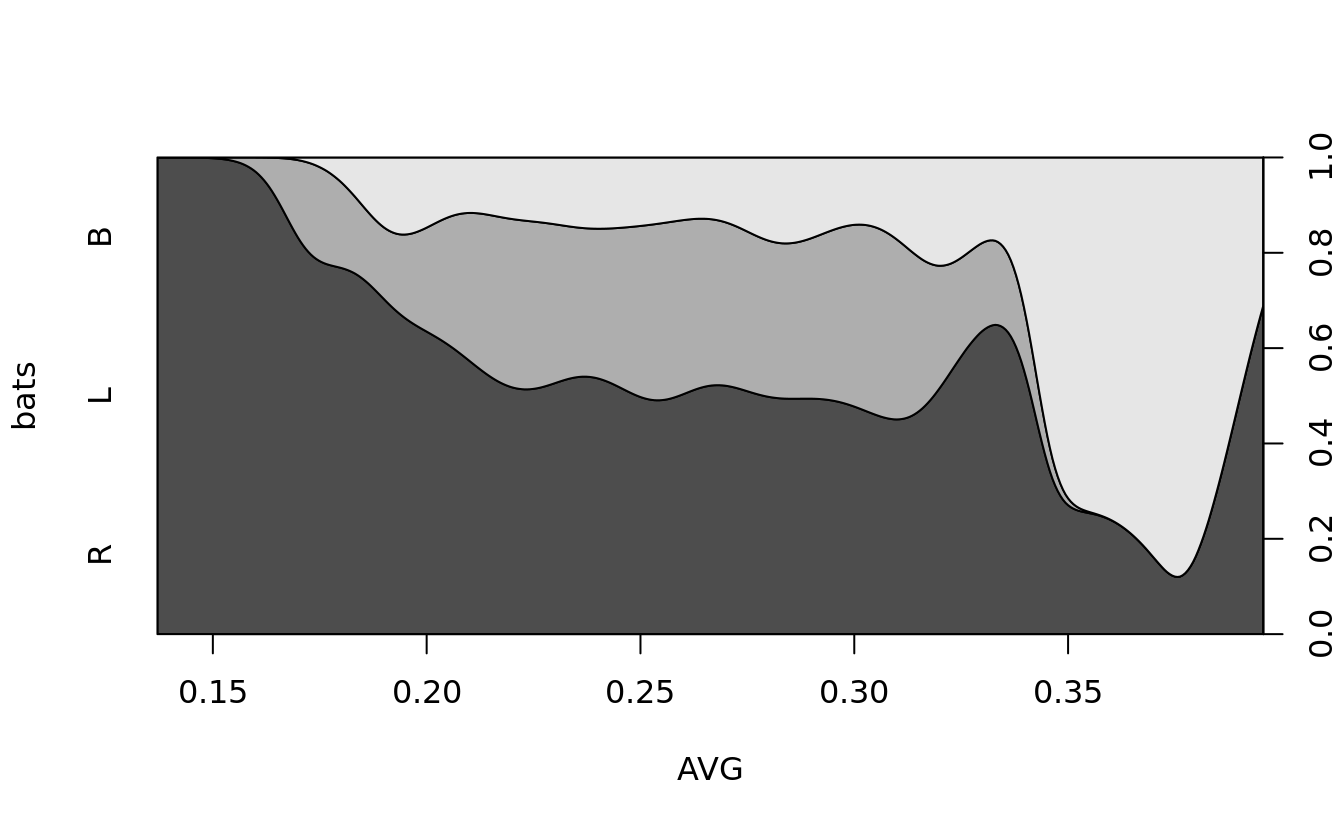
spineplot(bats~throws, bat.w.names)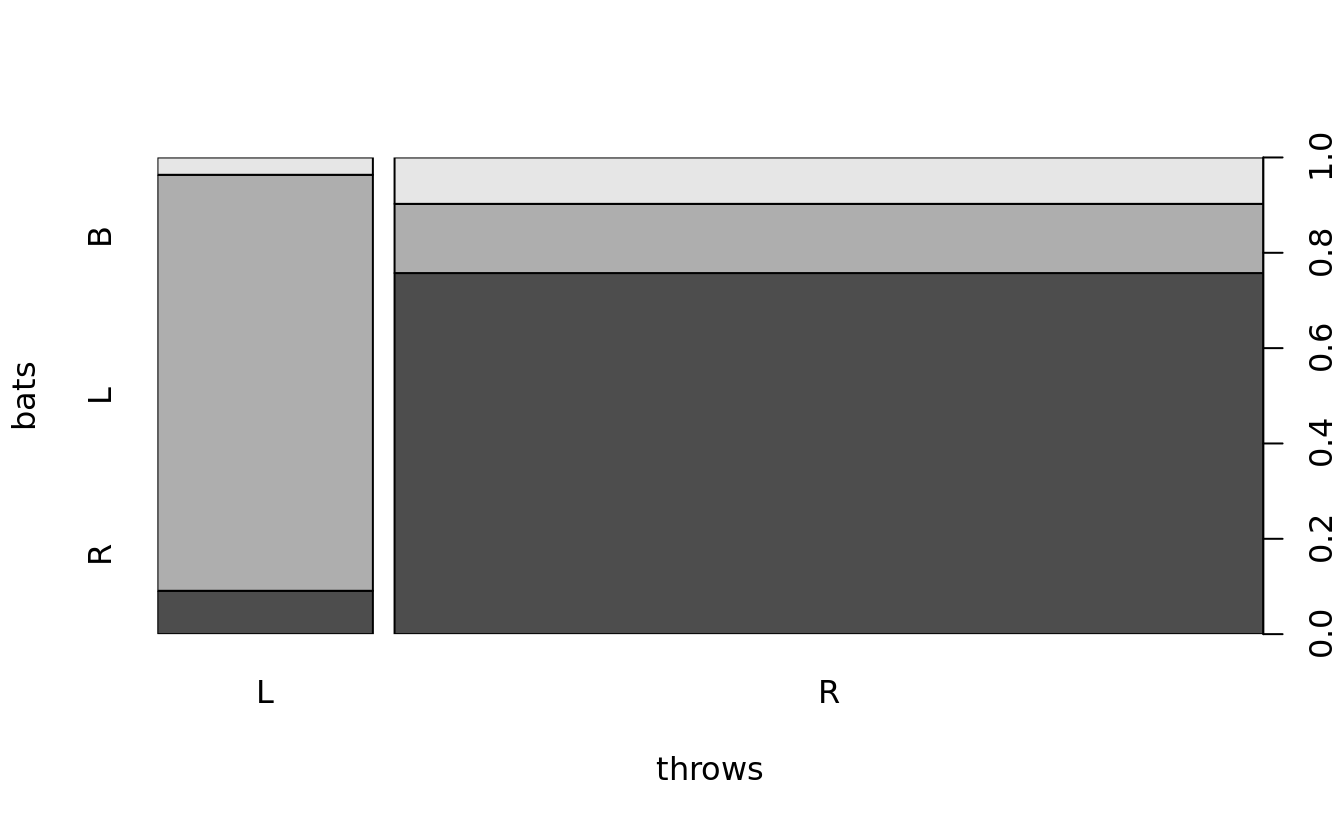
mosaicplot(bats~throws, bat.w.names, color = TRUE)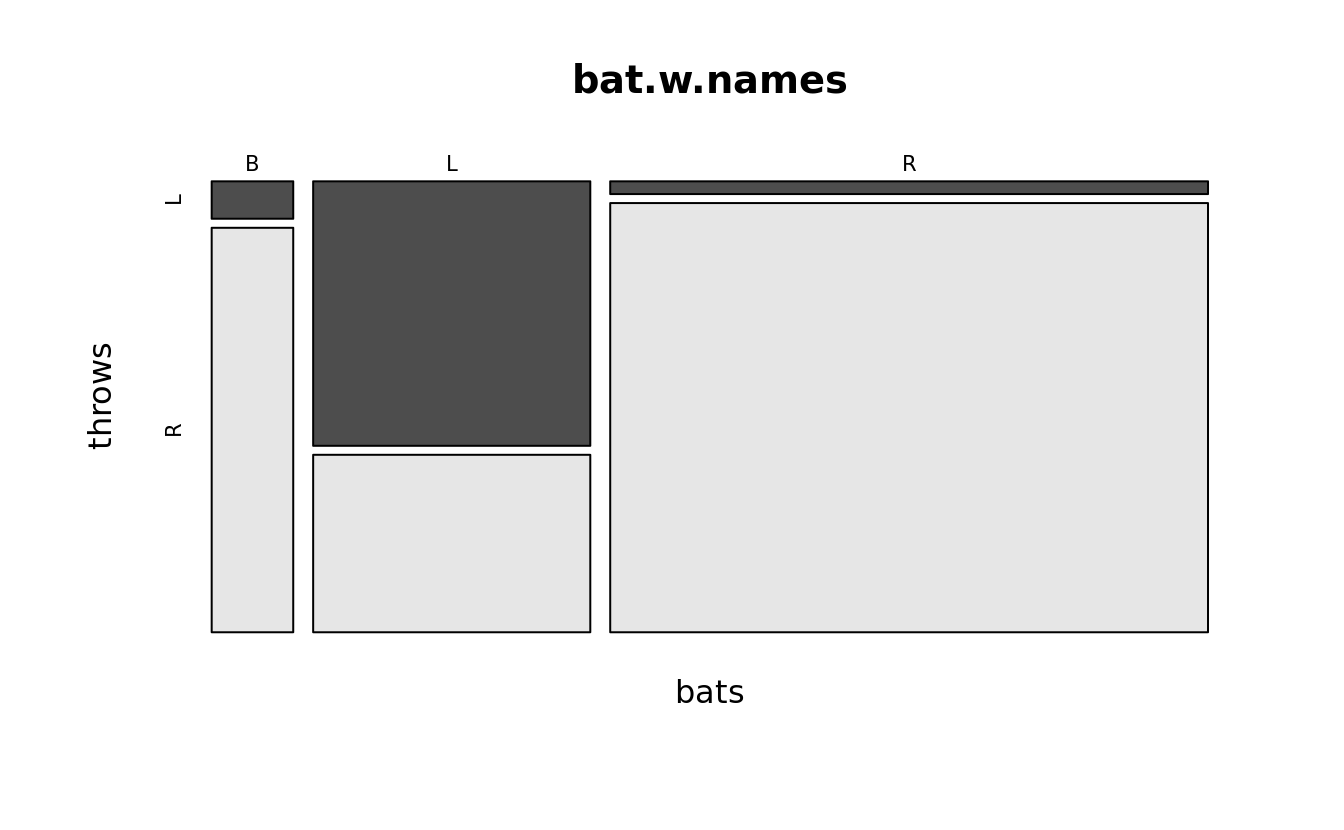
assocplot(table(bat.w.names$bats, bat.w.names$throws), xlab = "Throws", ylab = "Bats")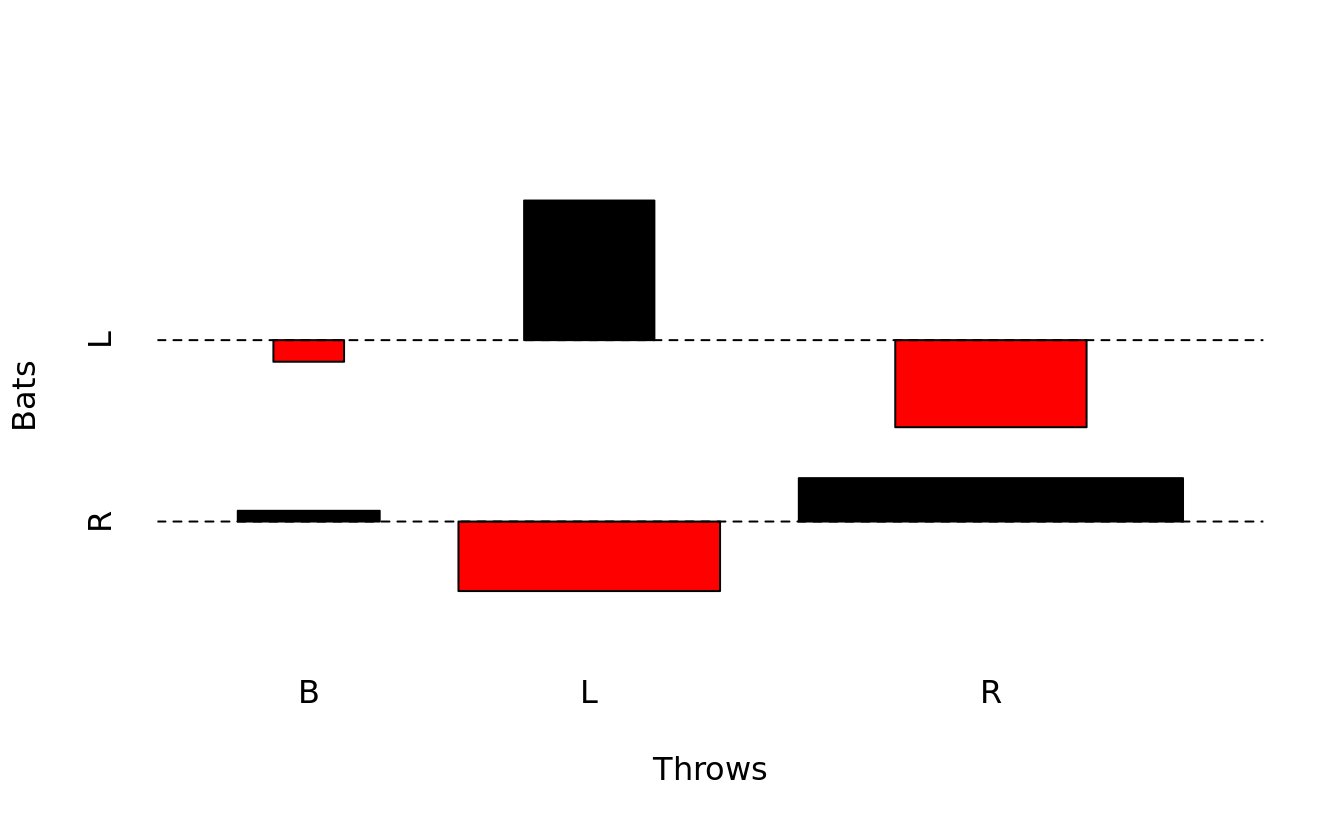
5.6 graphics: trivariate plot
we need to make two transformations:
- In the data file, values move east to west (or left to right) as x indices increase and from north to south (or top to bottom) as y indices increase. Unfortunately, persp plots increasing y coordinates from bottom to top.
- We need to select only a square subset of the elevation points.
To plot the figure, we rotate the image by 225?? (through theta=225), change the viewing angle to 20?? (phi=20), adjuste the light source to be from a 45?? angle (ltheta=45) and set the shading factor to 0.75 (shade=.75) to exaggerate topological features.
row = nrow(yosemite)
col = ncol(yosemite)
halfdome <- yosemite[(row - col + 1):row, col:1]5.6.1 persp() and headmap()
# three-dimensional surface for a specific perspective.
persp(halfdome, col = grey(.25), border = NA, expand = .15, theta = 225, phi = 20, ltheta = 45, lphi = 20, shade = .75)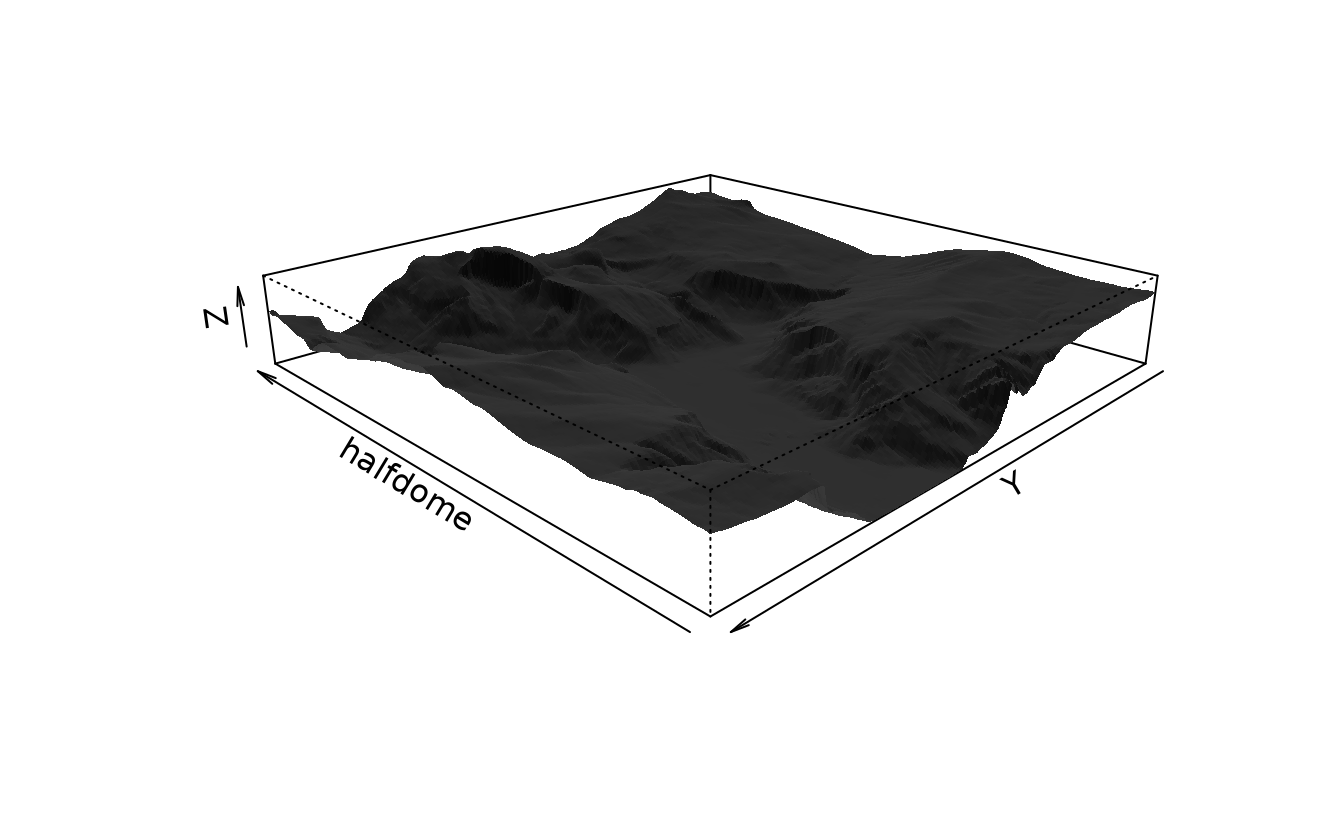
heatmap(yosemite)
5.6.2 image() and contour()
image() plot a matrix of data points as a grid of boxes, color coding the boxes based on the intensity at each location.
aspspecifies an aspect ratio that matches the dimensions of the data;
ylimspecifies that data is plotted from top to bottom
image(yosemite, asp = col/row, col = sapply((0:32)/32, gray))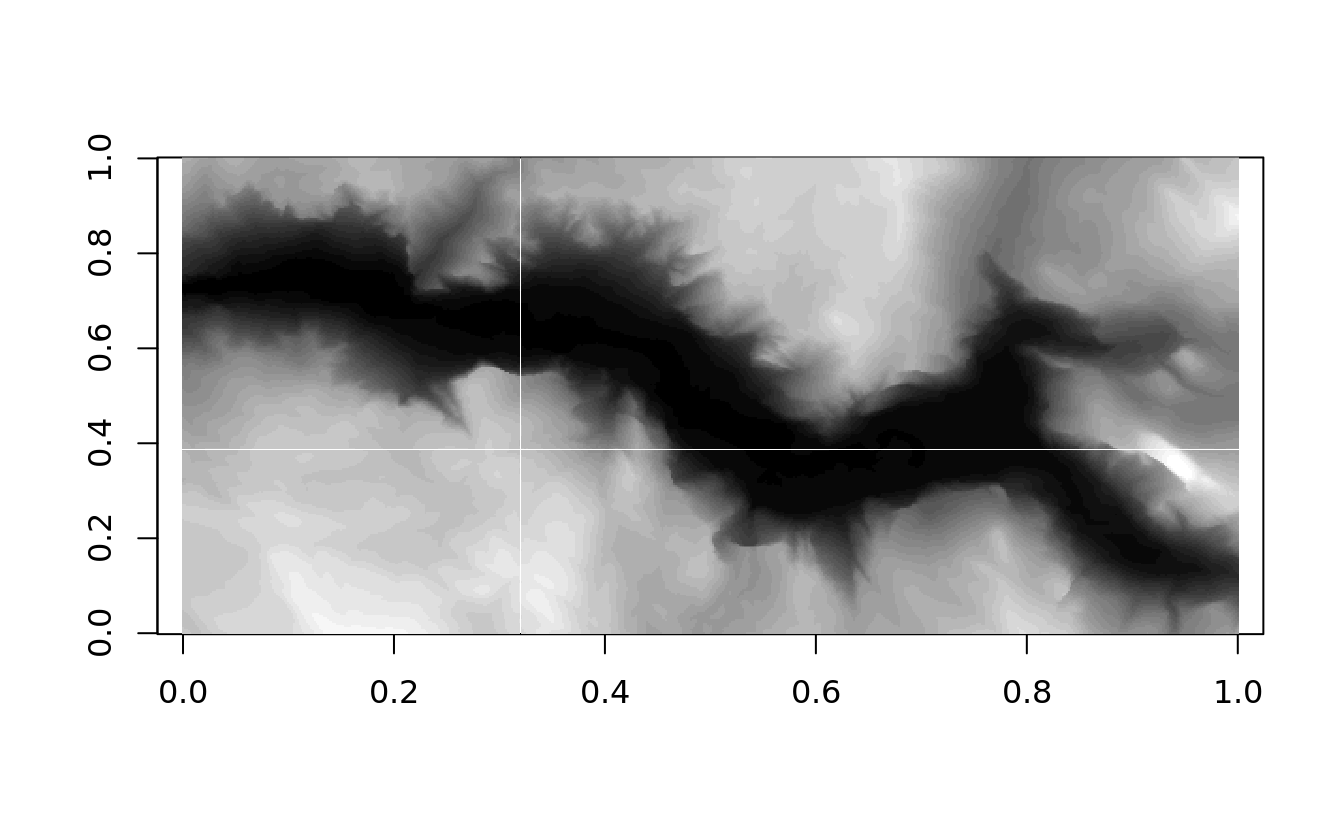
contour(yosemite, asp = col/row, ylim = c(1,0))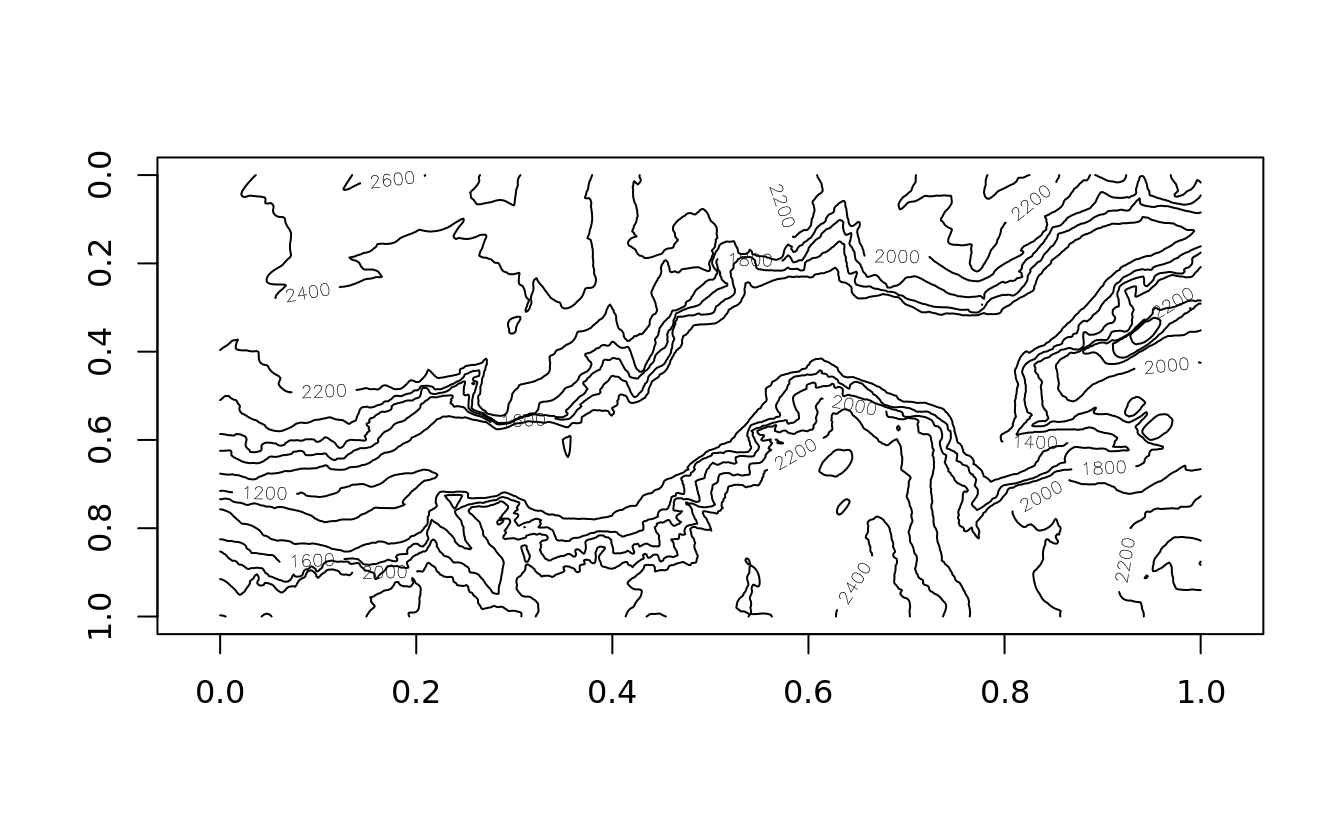
Contours are commonly added to existing image plots:
image(yosemite, asp = col/row, ylim = c(1,0), main = 'image + contour')
contour(yosemite, asp = col/row, ylim = c(1,0), add = T)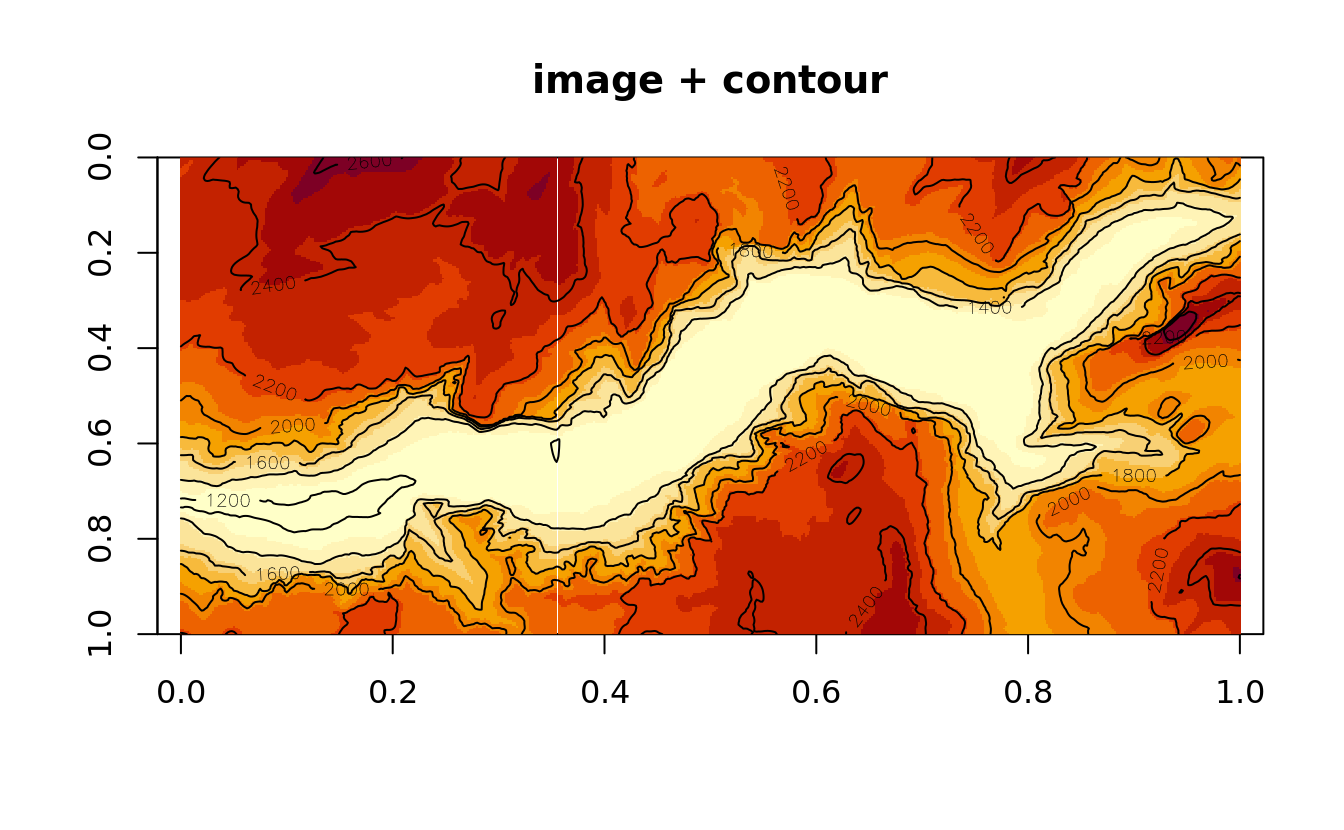
5.7 graphics: customizing charts
5.7.1 graphical parameters
- get
par("bg")
#> [1] "white"
head(par(),3)
#> $xlog
#> [1] FALSE
#>
#> $ylog
#> [1] FALSE
#>
#> $adj
#> [1] 0.5- set
par(bg = "transparent")5.7.2 mutliple plots
par(mfcol = c(2,2))
pie(5:1)
plot(1:400,rnorm(400))
qqnorm(rnorm(100))
hist(rnorm(10000))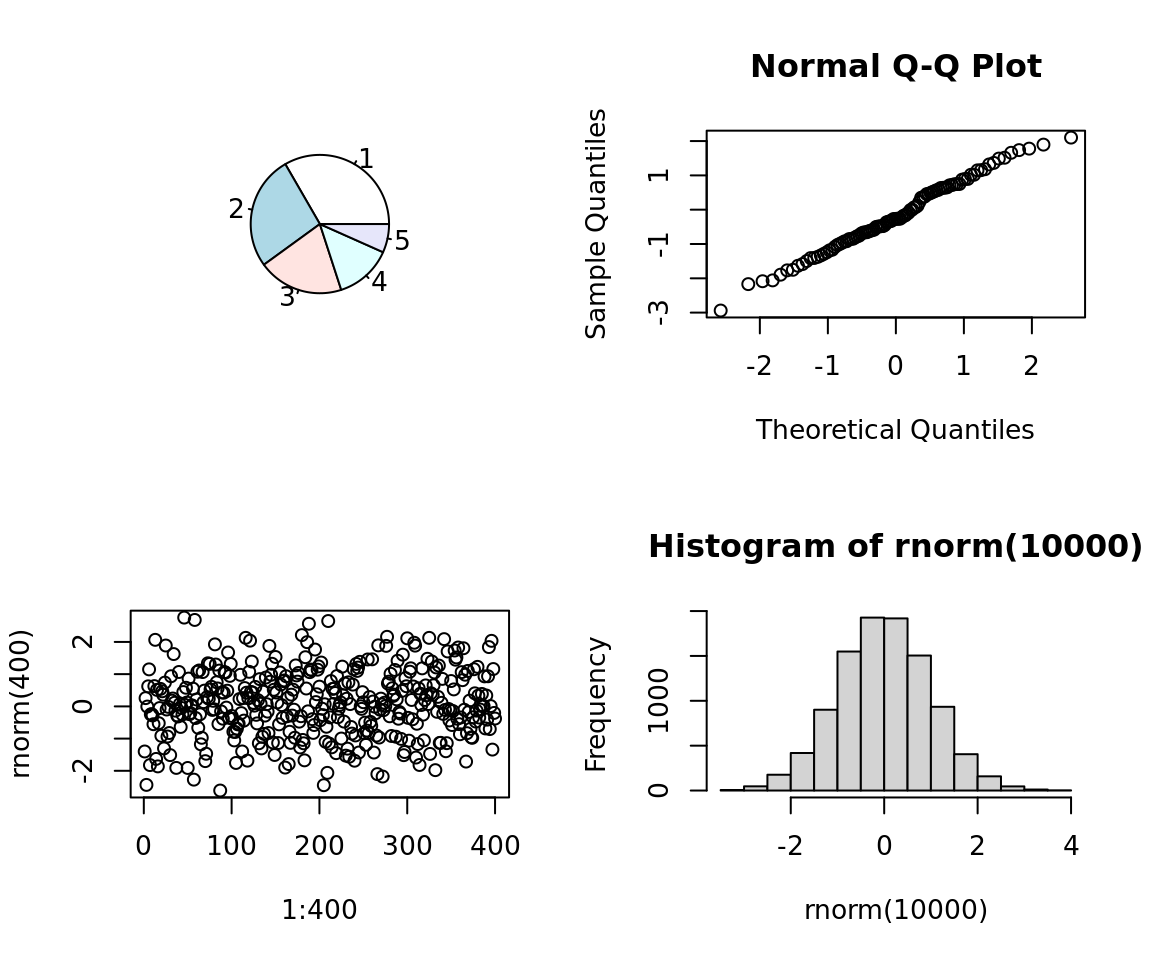
5.7.3 low-level graphics functions
abline()
plot(x = c(0,10),y = c(0,10), main = 'add lines')
abline(h = 4.5, col = 'red')
abline(v = 3.5, col = 'blue')
abline(a = 1, b = 1, col = 'green')
abline(coef = c(10,-1))
abline(h = 1:10,v = 1:10, col = 'gray')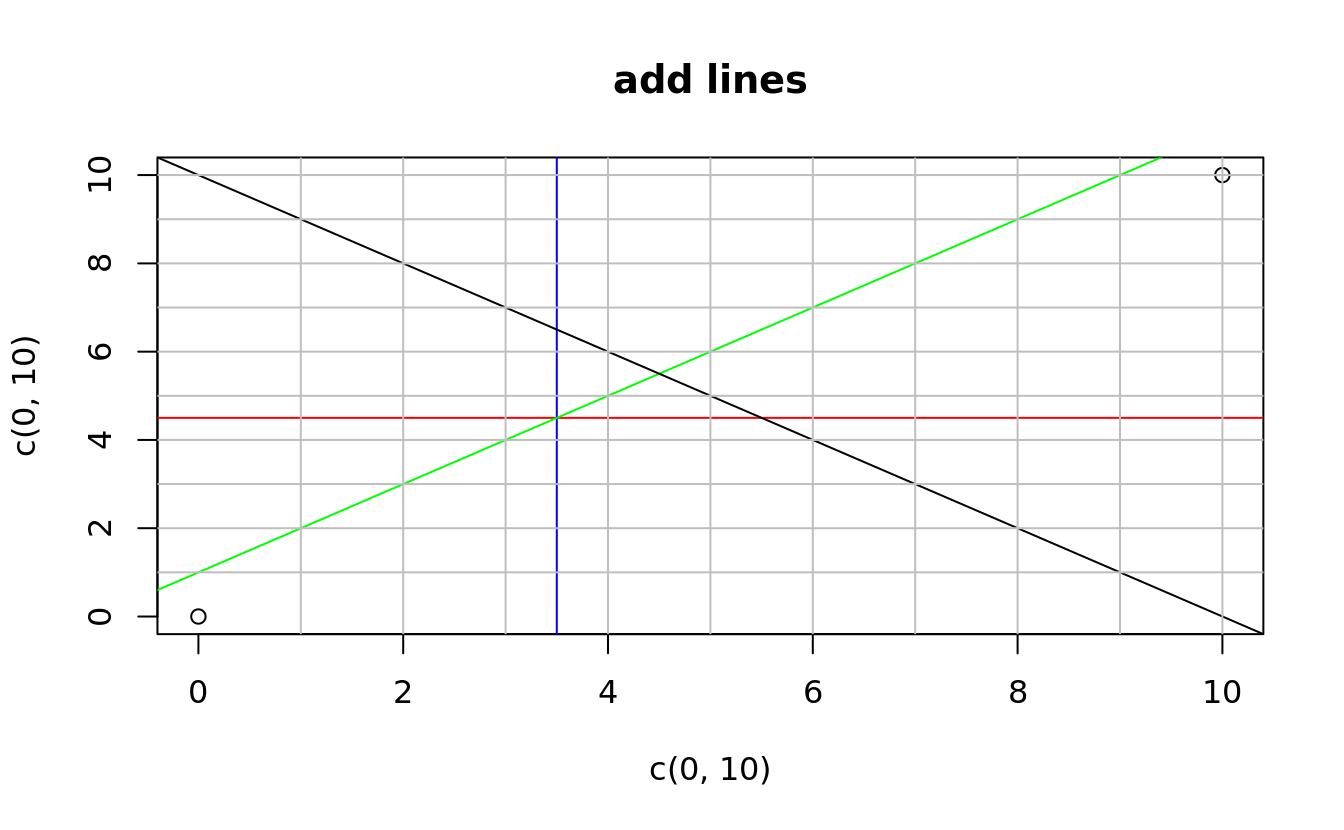
polygon()
plot(x = c(0,10),y = c(0,10), main = 'add polygon')
polygon(x = c(2,2,4,4),y = c(2,4,4,2))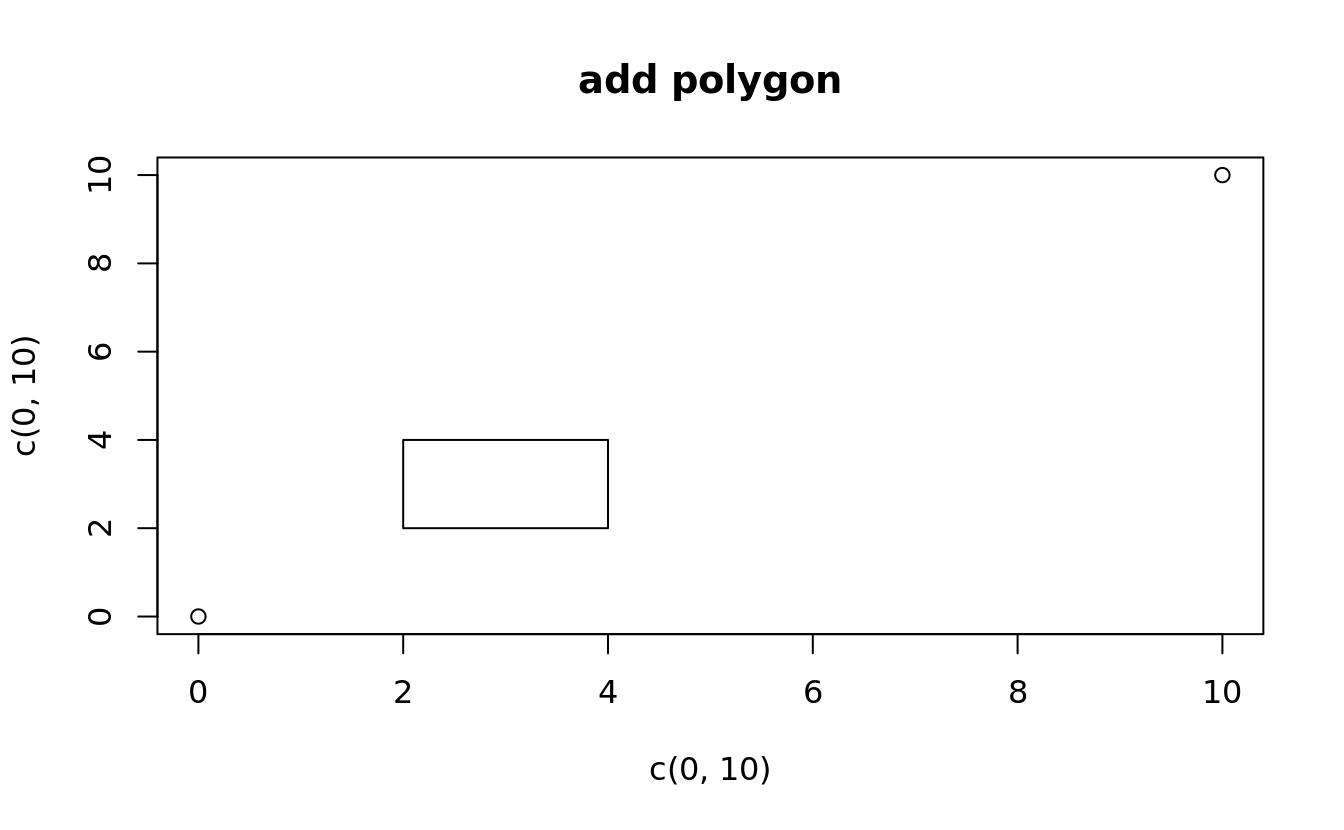
5.8 lattice: introduction
library(lattice)
library(nutshell)
library(learningr)
data(obama_vs_mccain, crab_tag)
data(births2006.smpl, tires.sus, sanfrancisco.home.sales, team.batting.00to08)
births = births2006.smpl
san = sanfrancisco.home.sales
obama_vs_mccain = obama_vs_mccain[!is.na(obama_vs_mccain$Turnout), ]
ovm = within(obama_vs_mccain, Region <- reorder(Region, Obama, median))
ovm2 <- ovm[!(ovm$State %in% c("Alaska", "Hawaii")), ]5.8.1 how lattice works
- The end user calls a high-level lattice plotting function.
- The lattice function examines the calling arguments and default parameters, assembles a lattice object, and returns the object. (Note that the class of the object is actually trellis.)
- The user calls plot.trellis or plot.trellis with the lattice object as an argument. (This typically happens automatically on the R console.)
- The function plot.trellis sets up the matrix of panels, assigns packets to different panels and then calls the panel function specified in the lattice object to draw the individual panels.
5.8.2 a simple example
(d <- data.frame(x = c(0:9), y = c(1:10), z = c(rep(c("a", "b"), times = 5))))
#> x y z
#> 1 0 1 a
#> 2 1 2 b
#> 3 2 3 a
#> 4 3 4 b
#> 5 4 5 a
#> 6 5 6 b
#> 7 6 7 a
#> 8 7 8 b
#> 9 8 9 a
#> 10 9 10 bxyplot(y~x, data = d)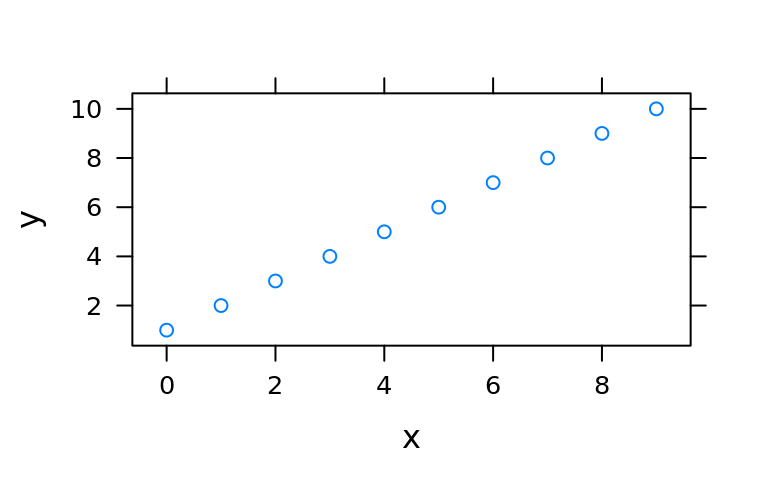
xyplot(y~x, groups = z, data = d)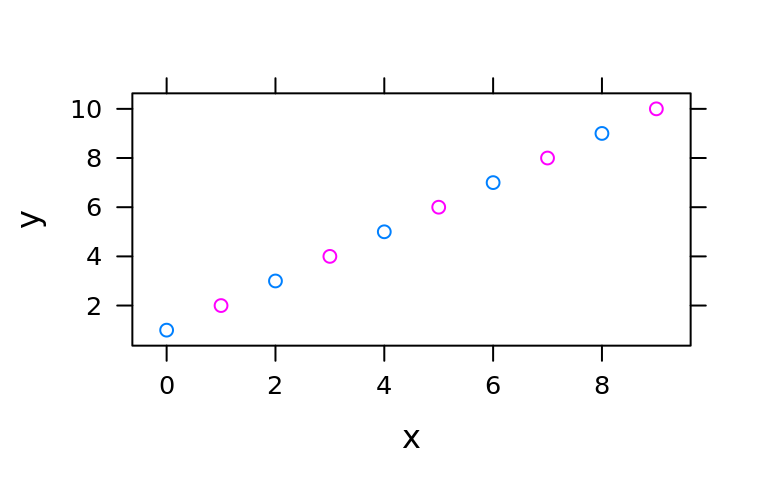
xyplot(y~x | z, data = d)
xyplot(
y~x | z, data = d,
panel = function(...){
panel.abline(a = 1,b = 1)
panel.xyplot(...)
}
)
5.9 lattice: univariate plot
5.9.1 bar chart
- one-dimentional table
(births.dow <- table(births$DOB_WK))
#>
#> 1 2 3 4 5 6 7
#> 40274 62757 69775 70290 70164 68380 45683
barchart(births.dow)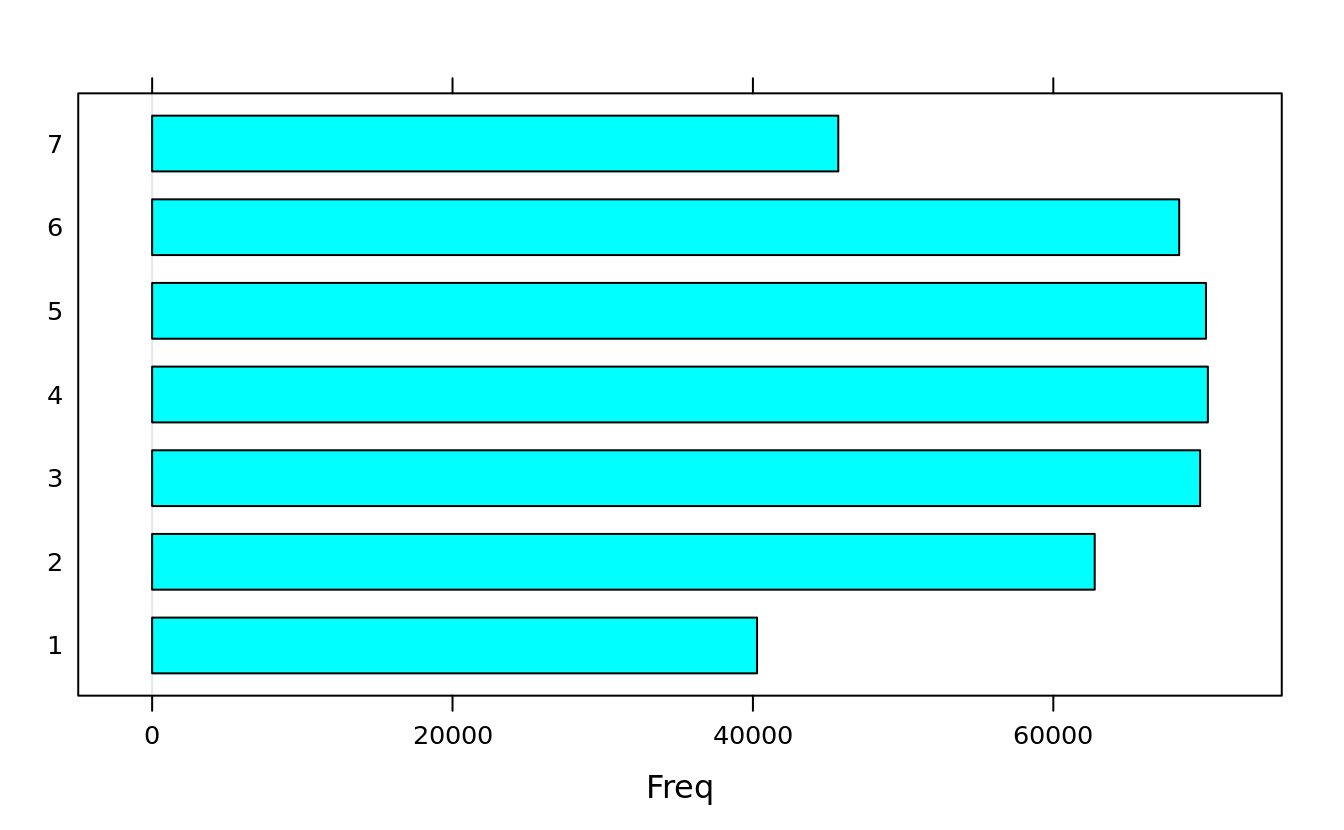
- two-dimensional table
births2006.dm <- plyr::mutate(births[births$DMETH_REC != "Unknown",],
DMETH_REC = as.factor(as.character(DMETH_REC)))
(dob.dm.tbl <- with(births2006.dm, table(WK = DOB_WK,MM = DMETH_REC)))
#> MM
#> WK C-section Vaginal
#> 1 8836 31348
#> 2 20454 42031
#> 3 22921 46607
#> 4 23103 46935
#> 5 22825 47081
#> 6 23233 44858
#> 7 10696 34878barchart(dob.dm.tbl)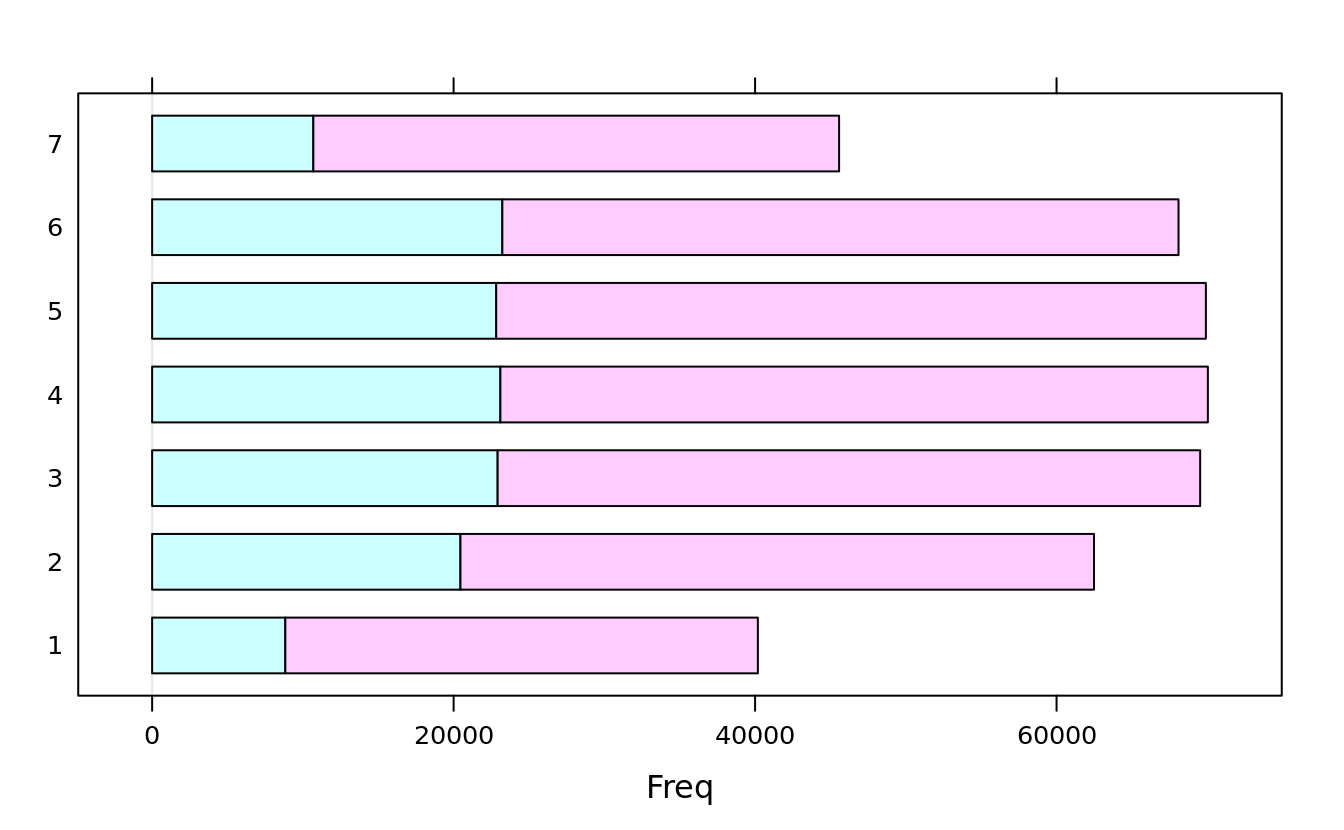
barchart(dob.dm.tbl, stack = F, auto.key = T)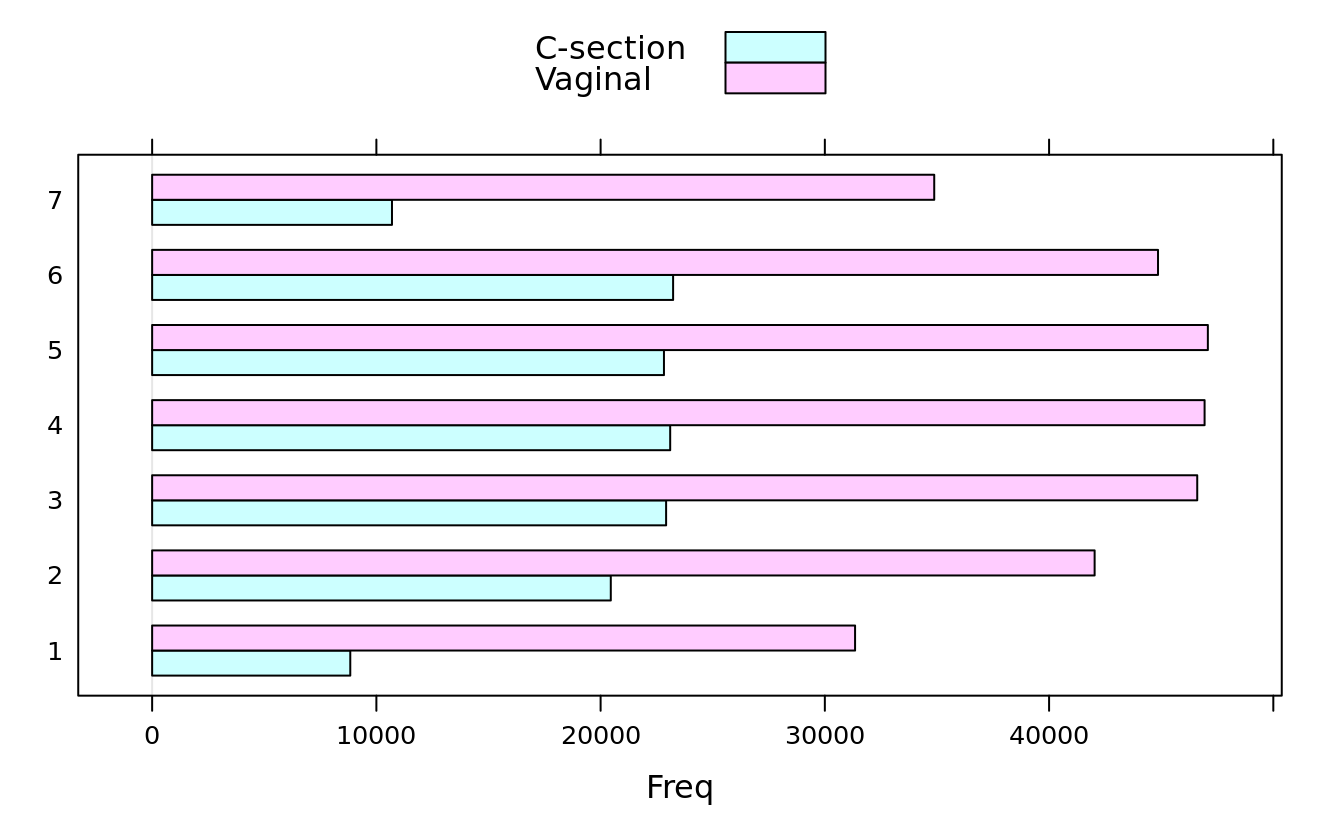
barchart(dob.dm.tbl, horizontal = F, groups = F)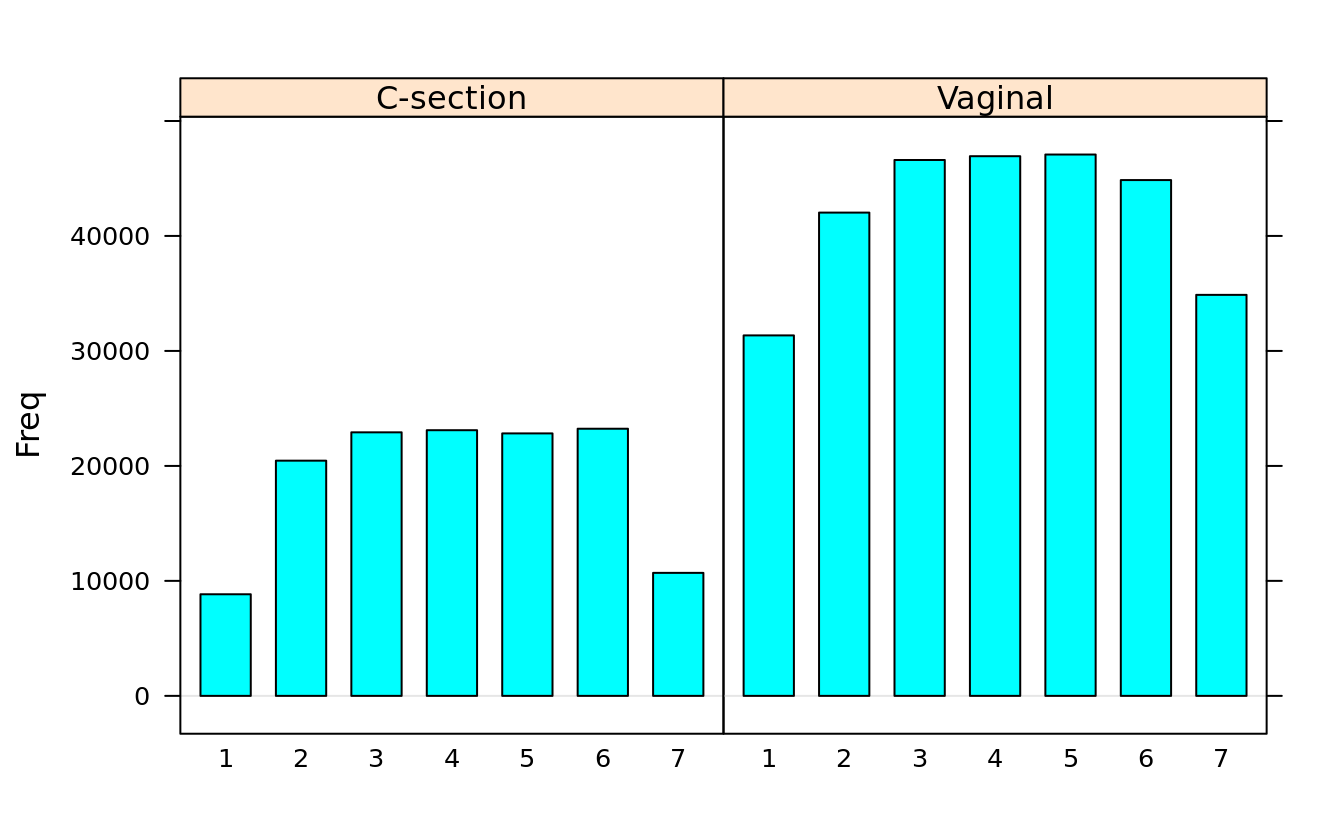
- data frame
ovm2 %>% tibble::as_tibble()
#> # A tibble: 45 x 15
#> State Region Obama McCain Turnout Unemployment Income Population Catholic
#> <fct> <fct> <dbl> <dbl> <dbl> <dbl> <int> <int> <int>
#> 1 Alab… IV 38.7 60.3 61.6 5.9 22732 4802982 6
#> 2 Ariz… IX 44.9 53.4 55.7 7.1 25203 6412700 29
#> 3 Arka… VI 38.9 58.7 53.1 5.8 20977 2926229 8
#> 4 Cali… IX 60.9 36.9 62 8.2 29020 37341989 37
#> 5 Colo… VIII 53.7 44.7 70.8 5.4 29679 5044930 21
#> 6 Dist… III 92.5 6.53 62 7.4 40846 601723 13
#> 7 Dela… III 61.9 36.9 66.7 5.9 28935 900877 26
#> 8 Flor… IV 50.9 48.1 67.4 7.3 26503 18900773 27
#> 9 Geor… IV 46.9 52.1 61.8 7.2 25098 9727566 9
#> 10 Idaho X 35.9 61.2 64.9 5.7 22262 1573499 10
#> # … with 35 more rows, and 6 more variables: Protestant <int>, Other <int>,
#> # Non.religious <int>, Black <dbl>, Latino <dbl>, Urbanization <dbl>barchart(State ~ Catholic, ovm2)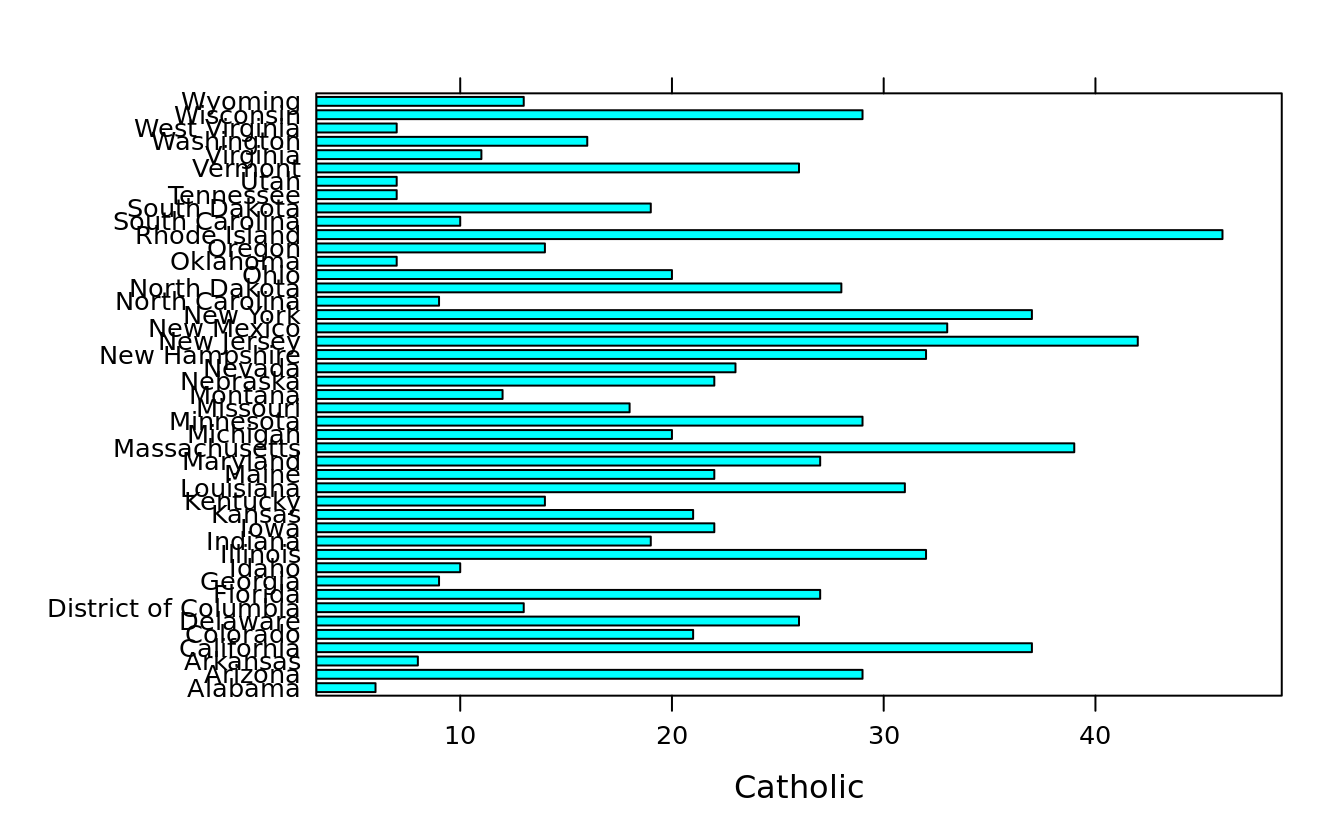
barchart(State ~ Catholic + Protestant + Non.religious + Other, ovm2, stack = T)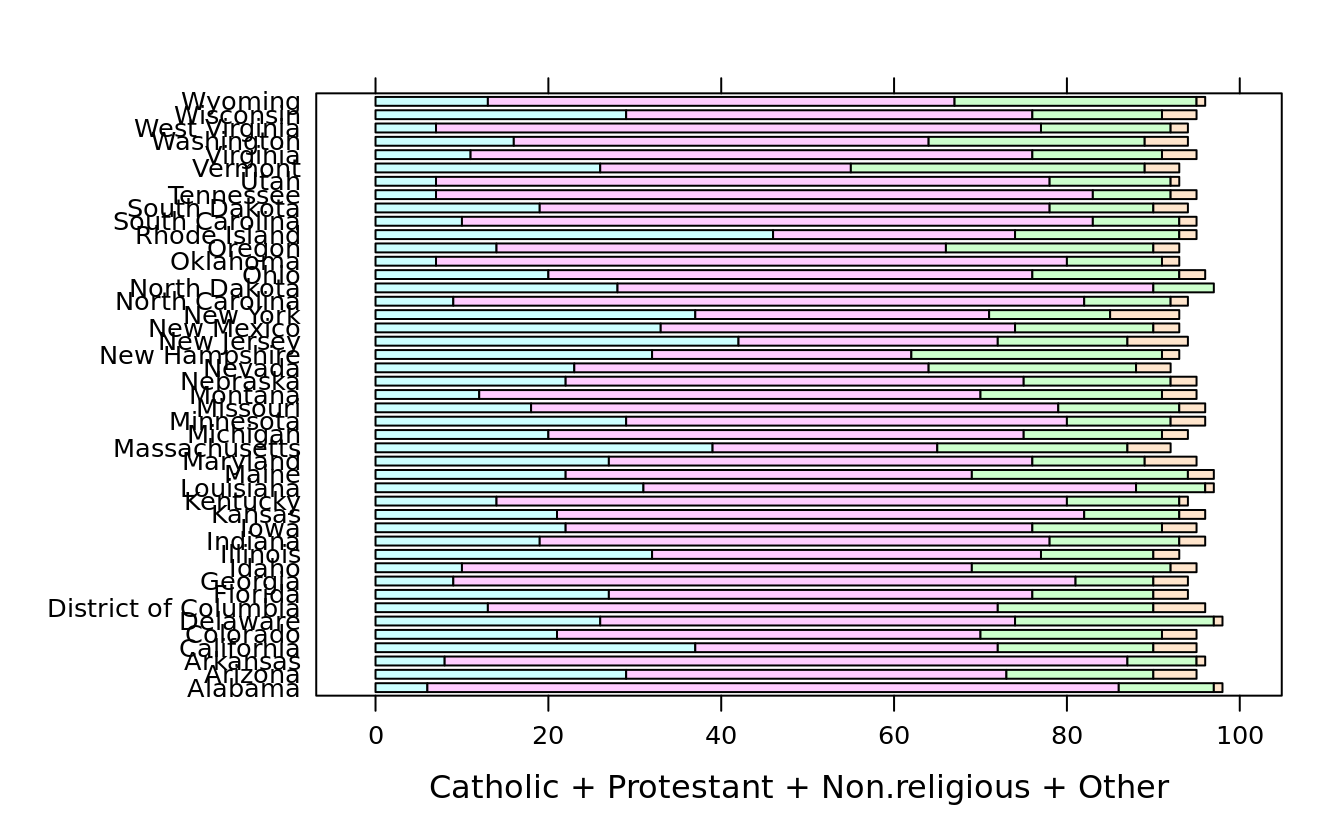
5.9.2 dot plot
- three-dimensional table
(dob.dm.tbl.alt <- with(births2006.dm, table(week = DOB_WK, month = DOB_MM, meths = DMETH_REC)))
#> , , meths = C-section
#>
#> month
#> week 1 2 3 4 5 6 7 8 9 10 11 12
#> 1 755 630 616 833 653 684 878 696 754 882 652 803
#> 2 1737 1475 1558 1641 1885 1667 2100 1691 1468 2049 1744 1439
#> 3 2044 1792 1686 1769 2090 1654 1589 2389 1936 2308 1844 1820
#> 4 1691 1738 2103 1678 2151 1860 1846 2306 1809 1779 2291 1851
#> 5 1663 1669 2114 1699 1787 2269 1872 2264 1891 1810 1946 1841
#> 6 1571 1767 2156 1678 1726 2292 1985 1825 2394 1821 1722 2296
#> 7 734 784 787 984 774 778 1123 853 1091 893 820 1075
#>
#> , , meths = Vaginal
#>
#> month
#> week 1 2 3 4 5 6 7 8 9 10 11 12
#> 1 2881 2289 2341 2778 2313 2341 3093 2459 2511 3072 2341 2929
#> 2 3892 3239 3205 3192 3797 3341 4134 3414 3361 4102 3270 3084
#> 3 4196 3490 3546 3509 4355 3513 3542 4807 3847 4563 3750 3489
#> 4 3425 3525 4359 3467 4324 3665 3634 4679 3900 3646 4522 3789
#> 5 3454 3578 4433 3445 3541 4567 3892 4651 3915 3707 4200 3698
#> 6 3236 3517 4152 3337 3231 4298 3756 3684 4598 3422 3328 4299
#> 7 2550 2601 2611 3191 2580 2682 3616 2824 3565 2663 2684 3311
dotplot(dob.dm.tbl.alt, groups = T, auto.key = T, stack = F)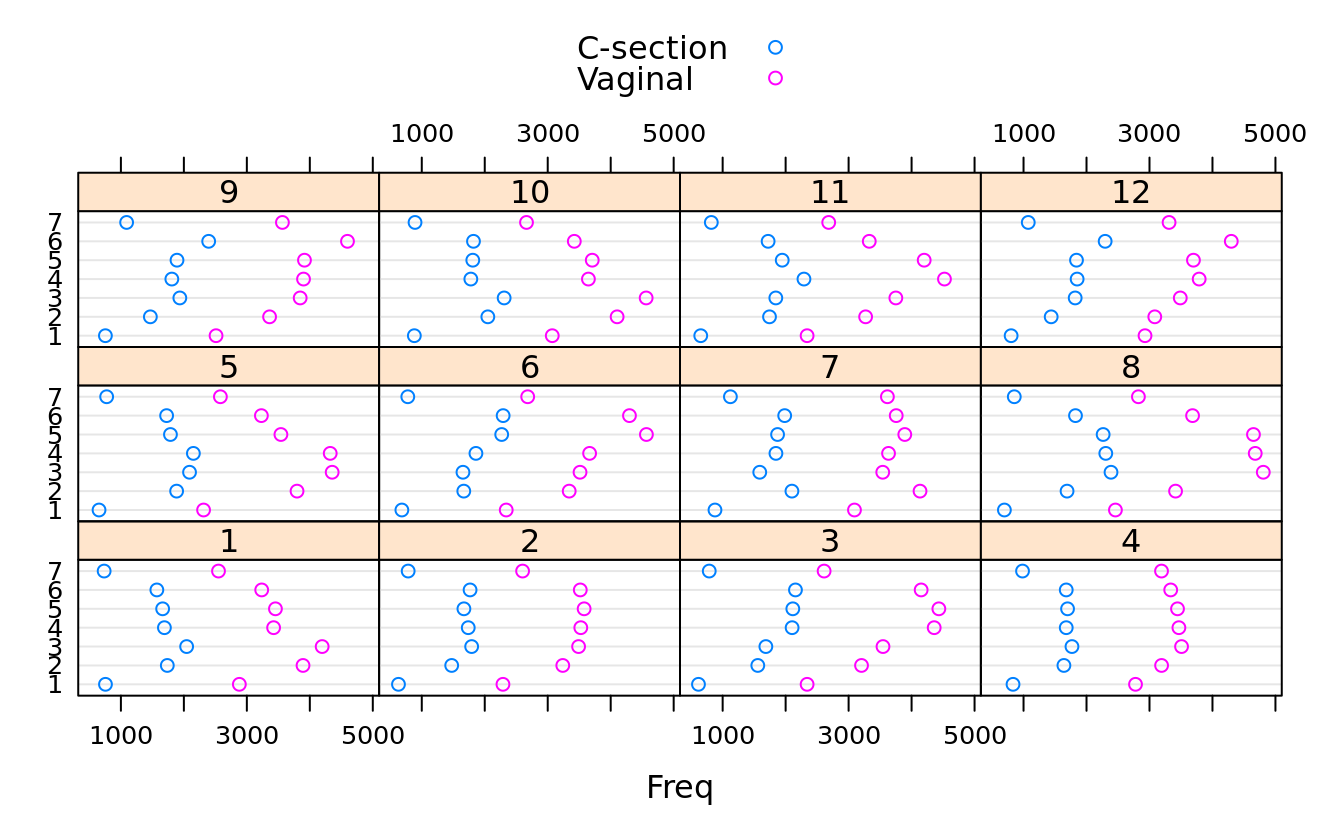
- data frame
tires.sus$Speed_At_Failure_km_h %>% tibble::as_tibble()
#> # A tibble: 66 x 1
#> value
#> <int>
#> 1 180
#> 2 180
#> 3 180
#> 4 180
#> 5 180
#> 6 180
#> 7 180
#> 8 180
#> 9 180
#> 10 180
#> # … with 56 more rows
dotplot(as.factor(Speed_At_Failure_km_h) ~ Time_To_Failure | Tire_Type, tires.sus)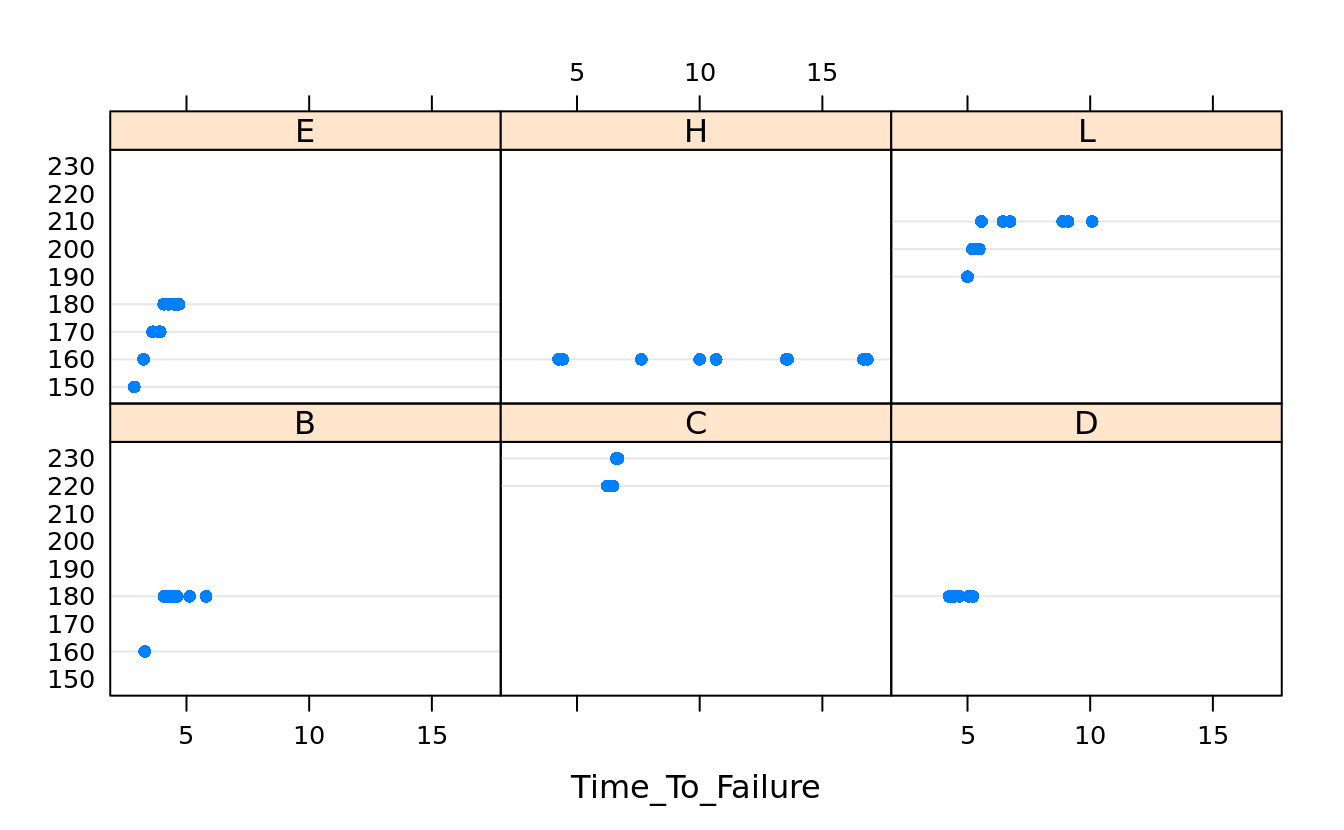
5.9.3 histogram
histogram(~ Obama, obama_vs_mccain) 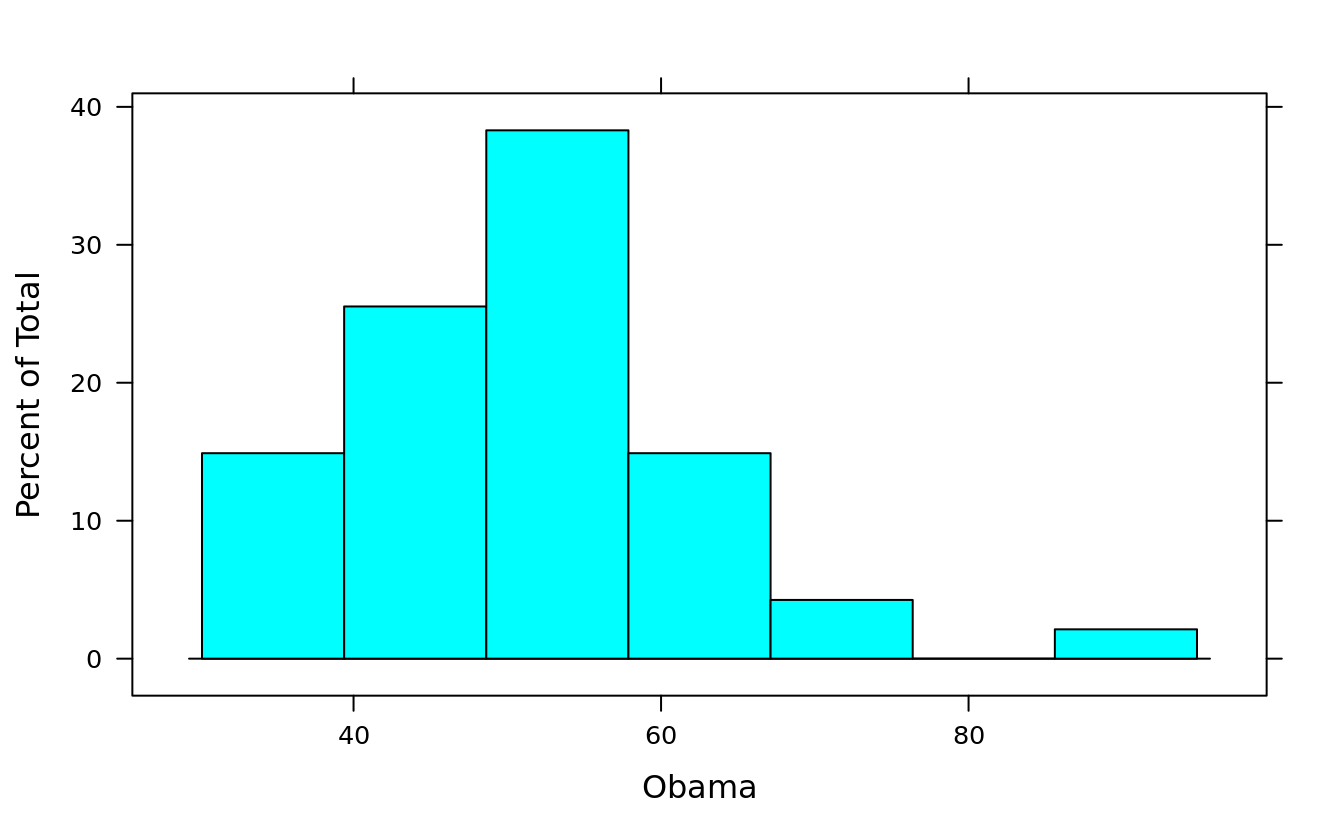
histogram(~ Obama, obama_vs_mccain, breaks = 10)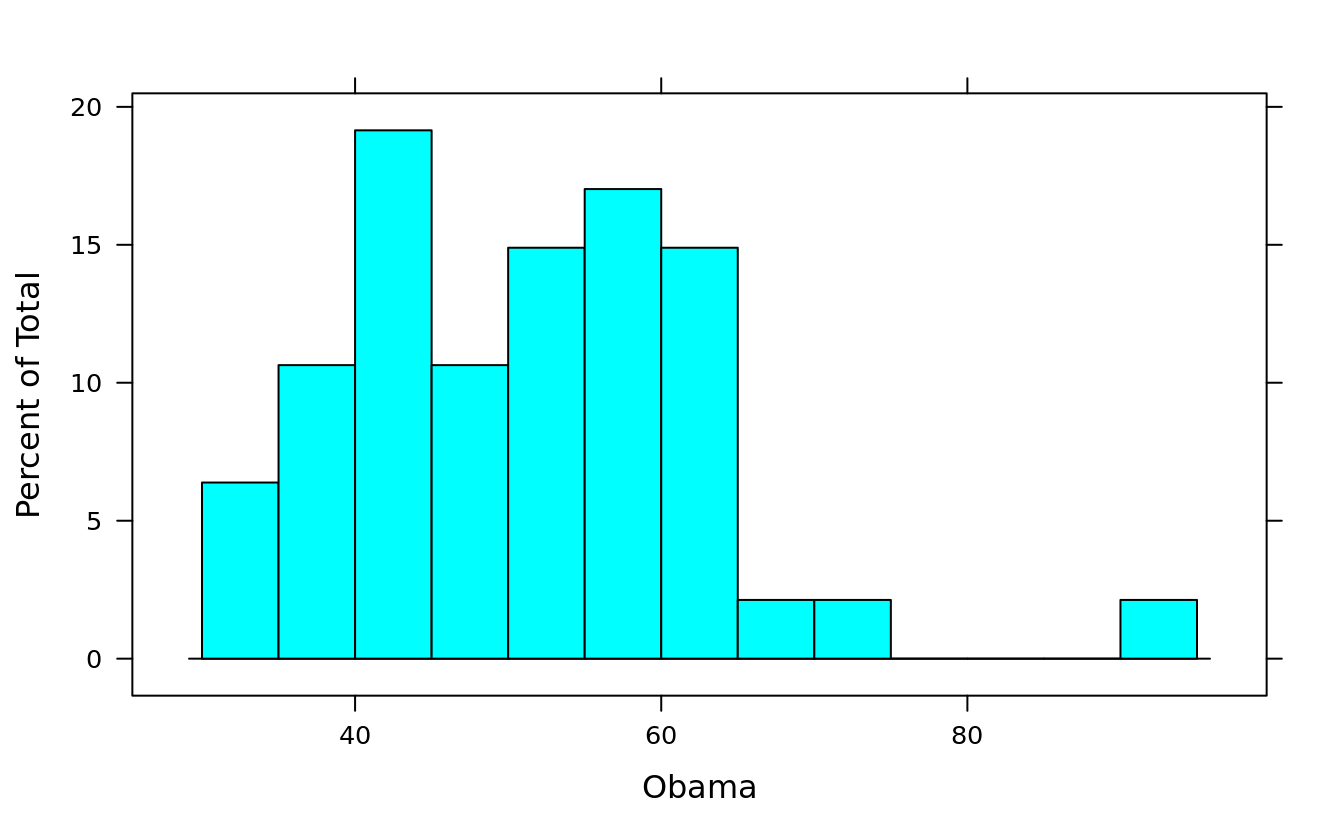
histogram(~ Obama, obama_vs_mccain, type = "percent")
histogram(~ Obama, obama_vs_mccain, type = "count")
histogram(~ Obama, obama_vs_mccain, type = "density")
histogram(~DBWT | DPLURAL, births)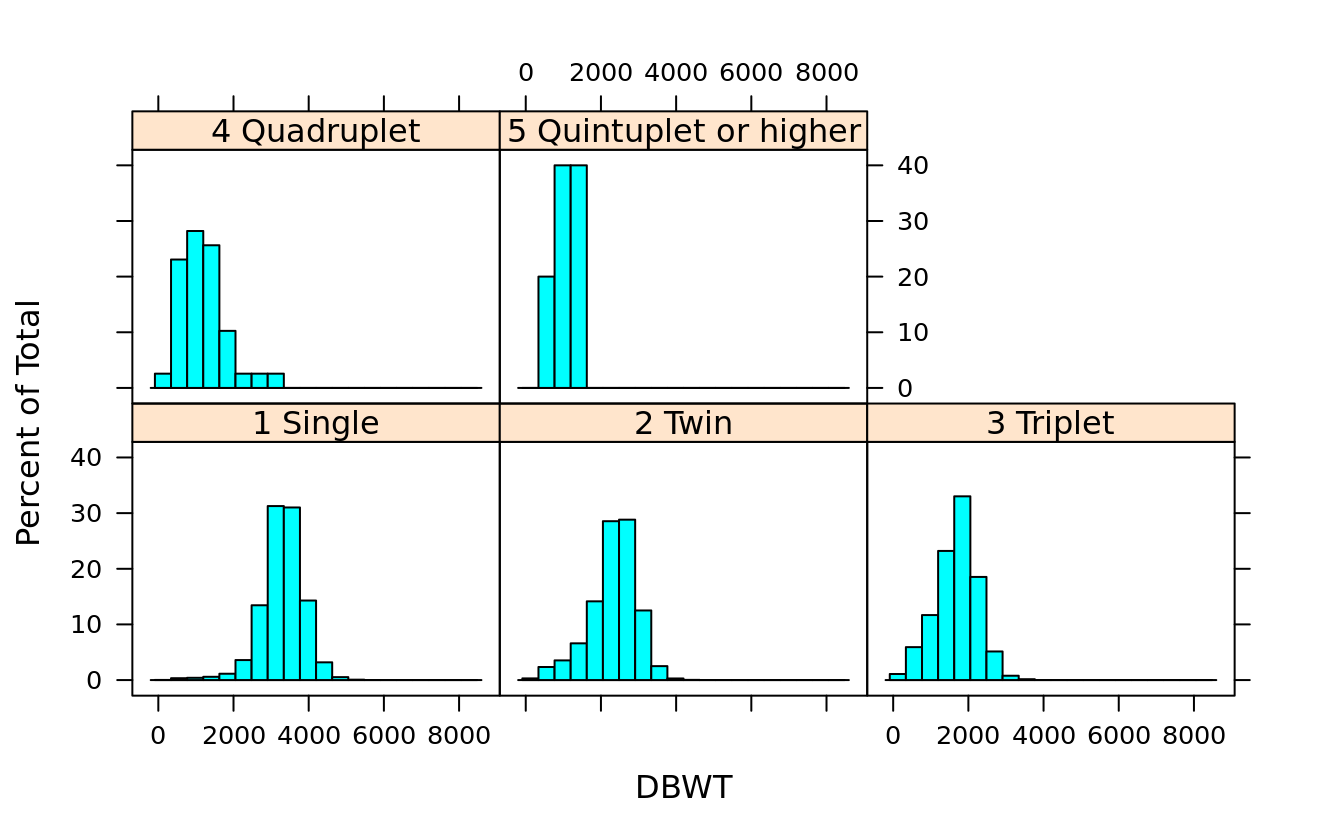
histogram(~DBWT | DPLURAL, births, layout = c(1,5))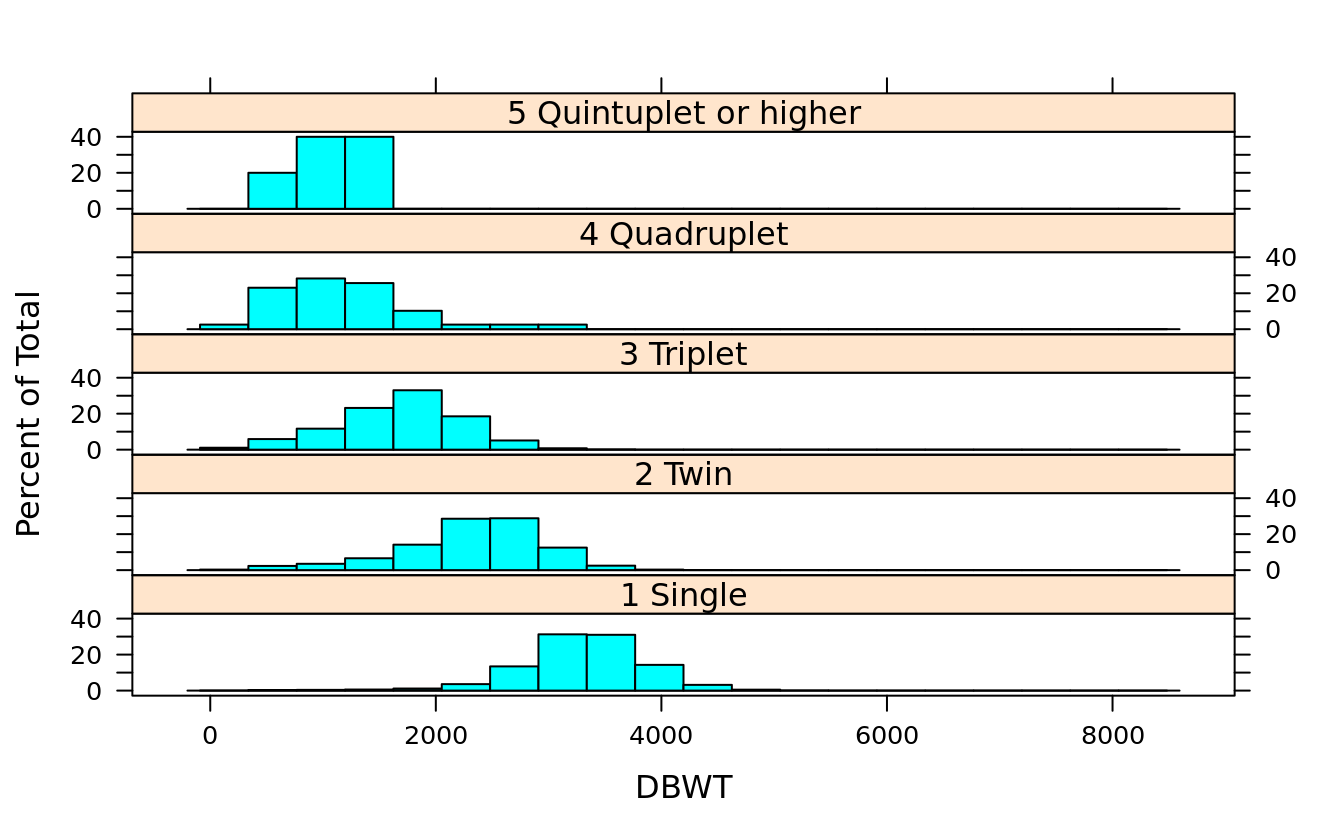
5.9.4 denstity plot
(plur = unique(as.character(births$DPLURAL))[4:5]) #quadruplet or higher
#> [1] "4 Quadruplet" "5 Quintuplet or higher"#By default, densityplot will draw a strip chart under each chart:
densityplot(~DBWT | DPLURAL, births, subset = (DPLURAL %in% plur))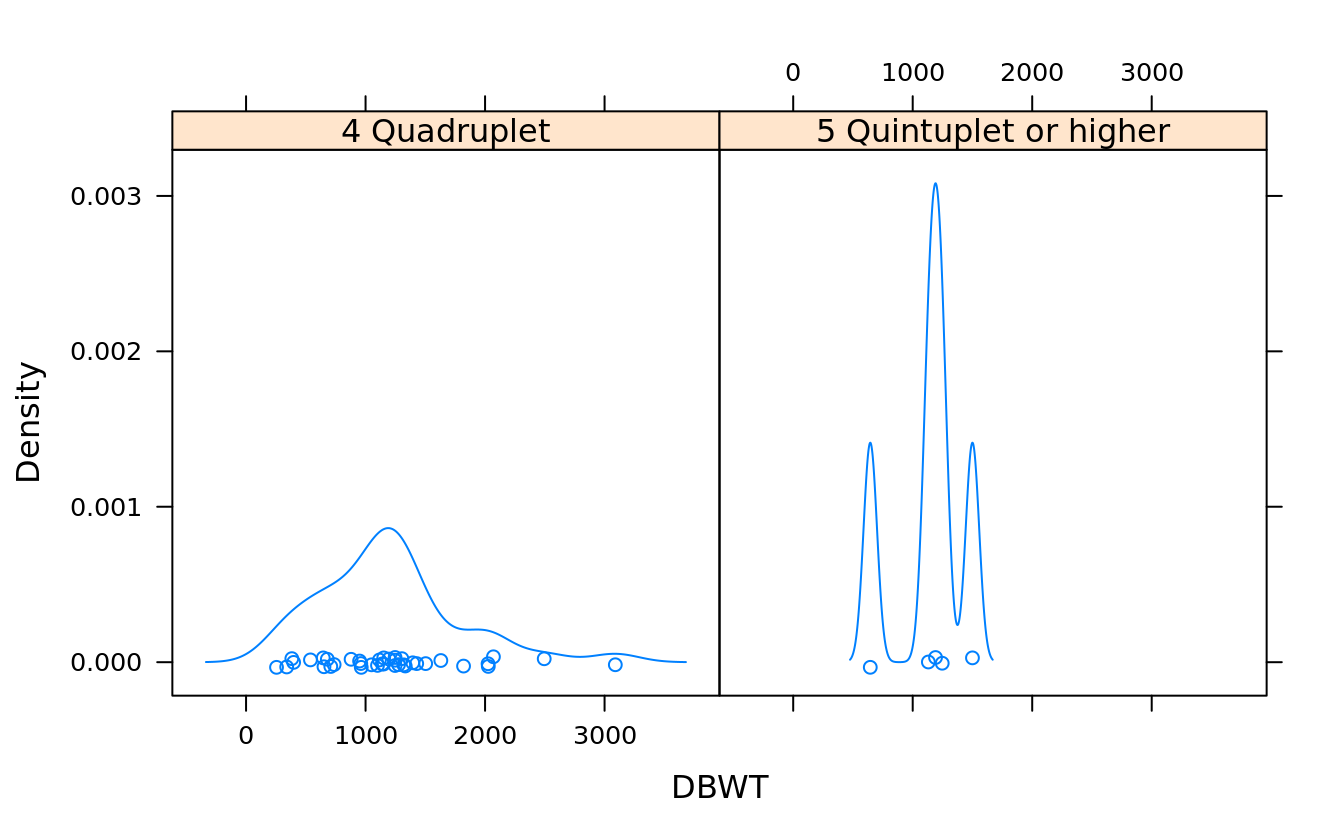
#turn off the strip by plot.points = F because the data set is so big
densityplot(~DBWT | DPLURAL, births, layout = c(1,5), plot.points = F)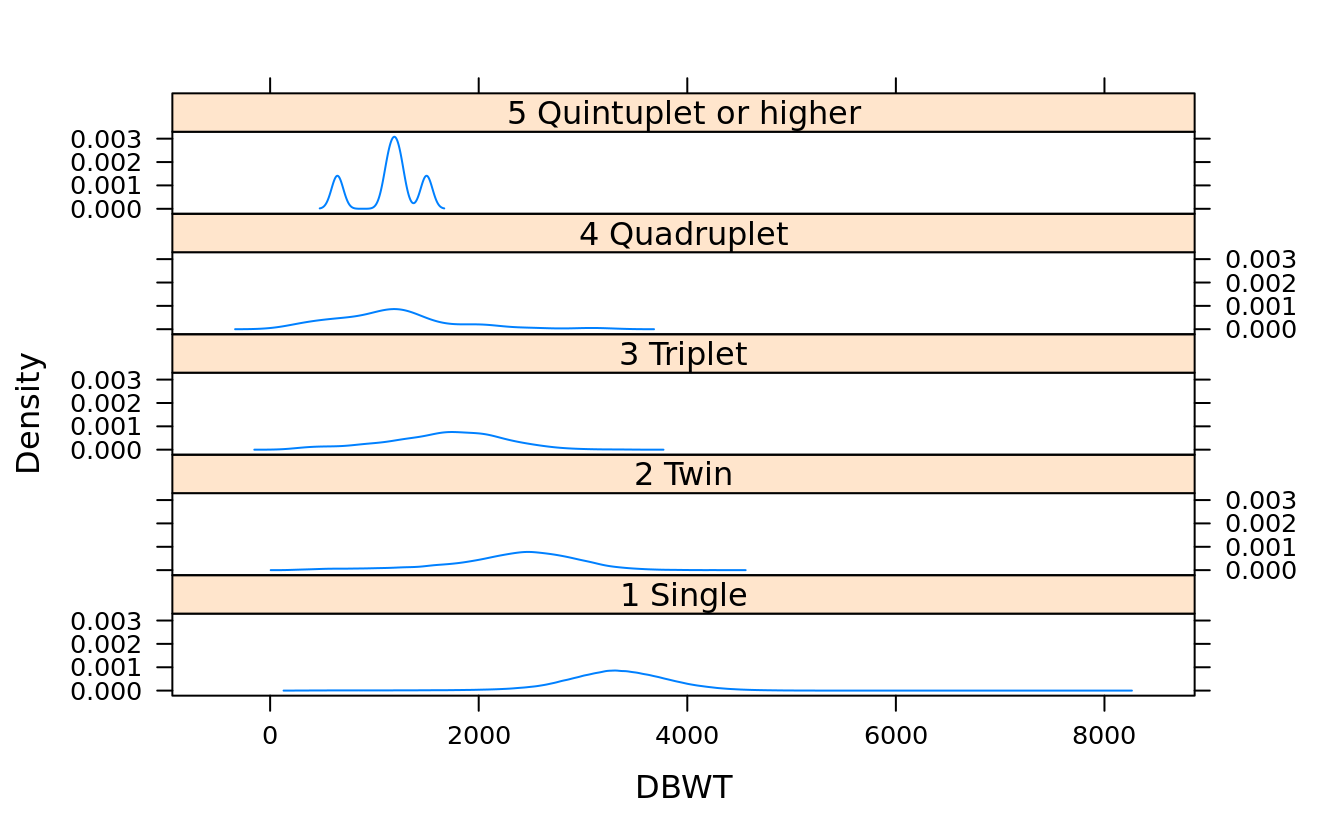
densityplot(~DBWT, groups = DPLURAL, births, plot.points = F, auto.key = T)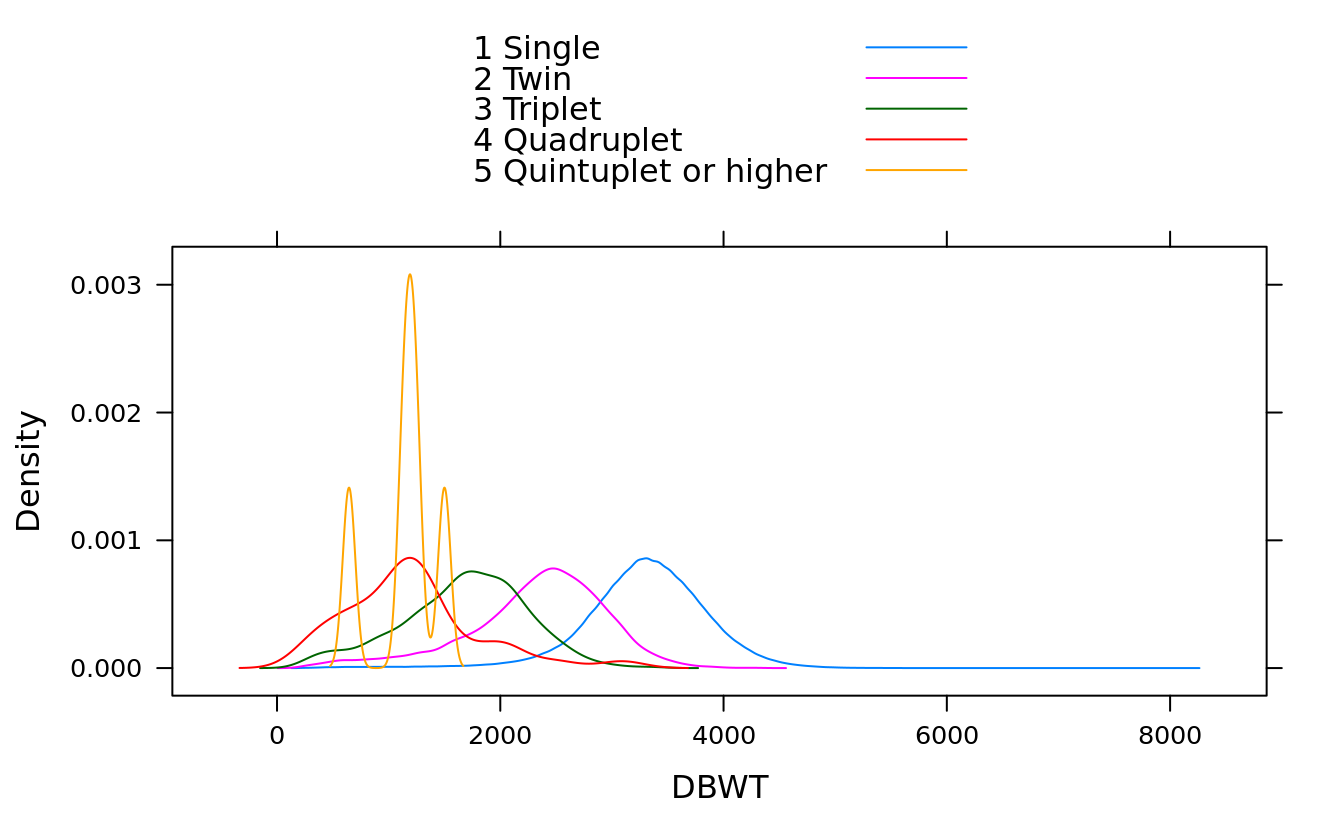
5.9.5 strip plot
You can think of strip plots as one-dimensional scatter plots:
#jitter.data=T adds some random vertical noise to make the points easier to read
stripplot(~DBWT, births, jitter.data = T, subset = (DPLURAL %in% plur))
5.9.6 quantile plot
qqmath(rnorm(100000))
qqmath(~DBWT | DPLURAL, births, pch = 19, cex = .25)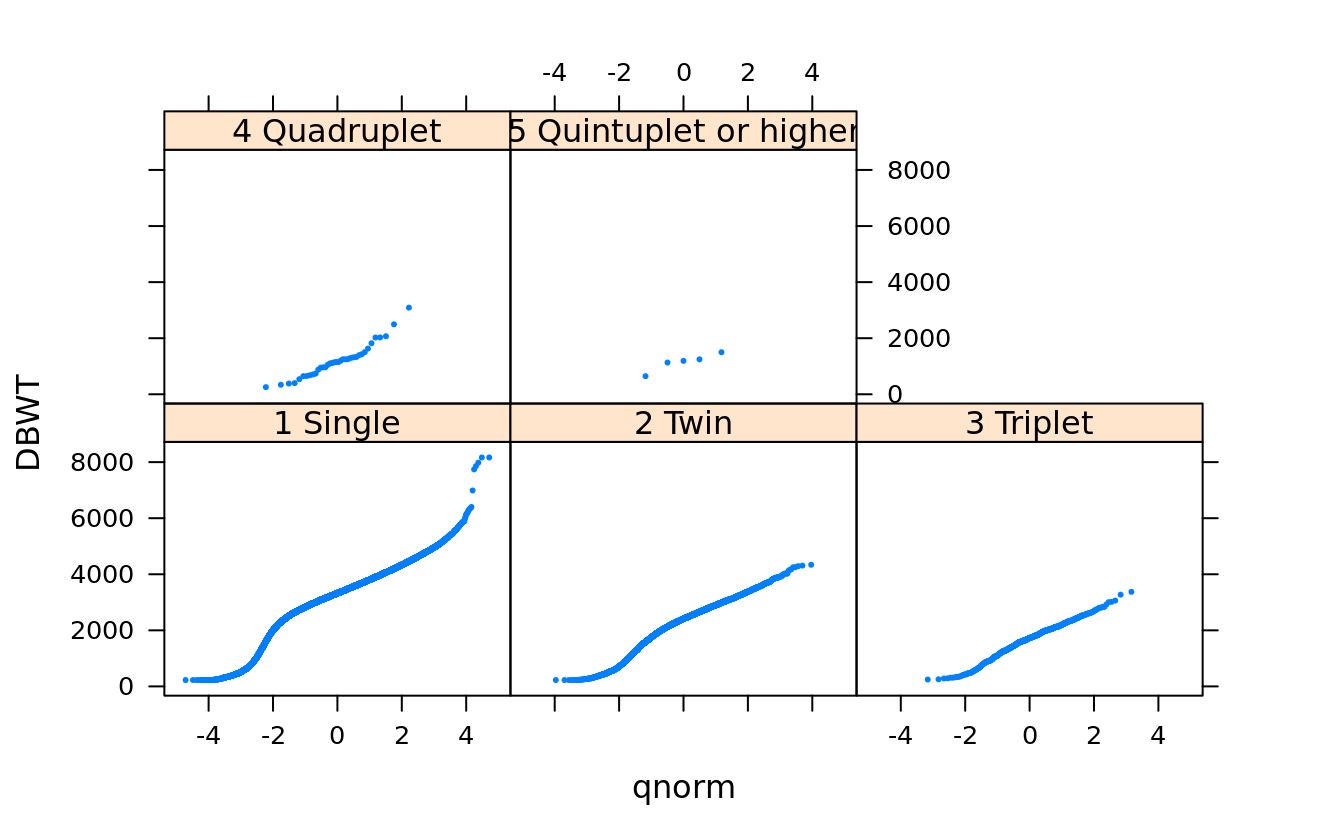
qqmath(~price, san)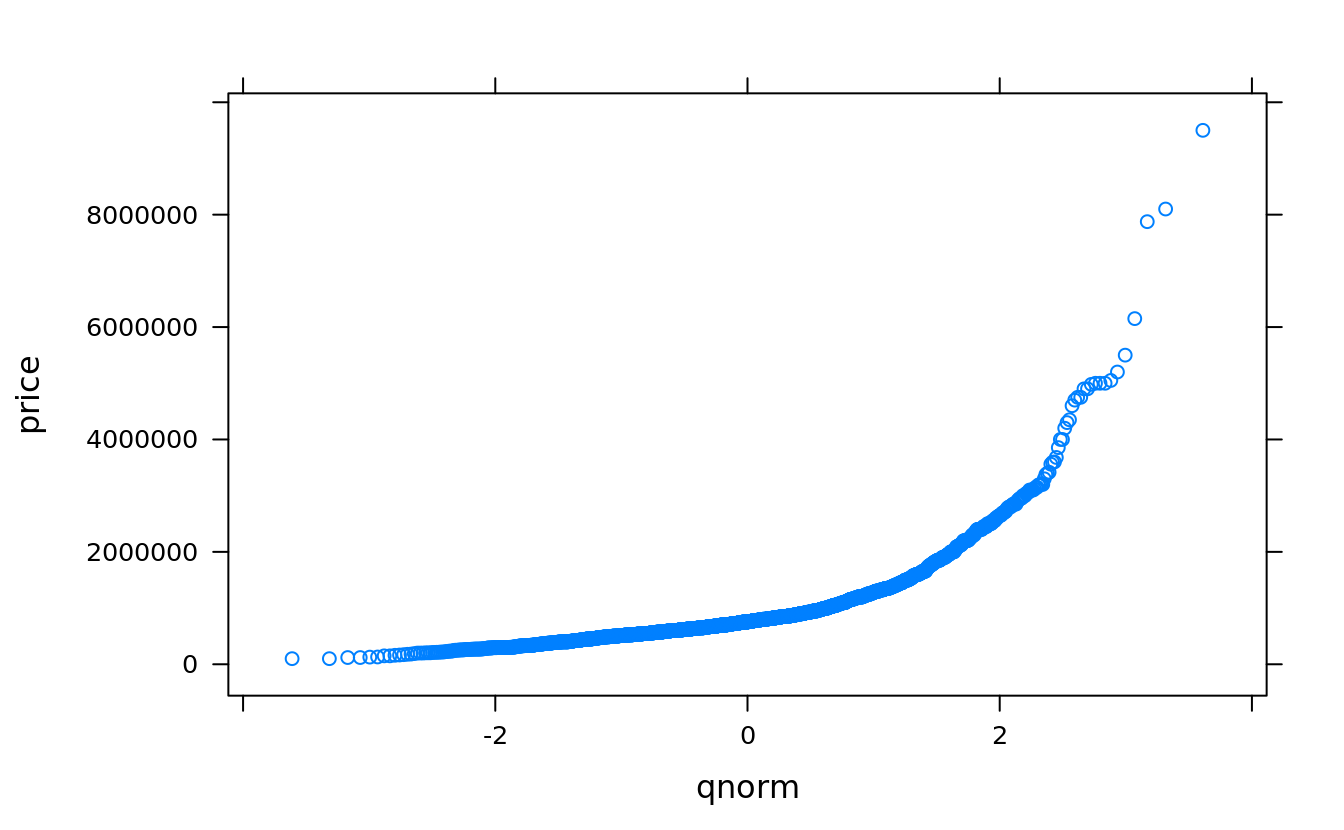
qqmath(~log(price), san)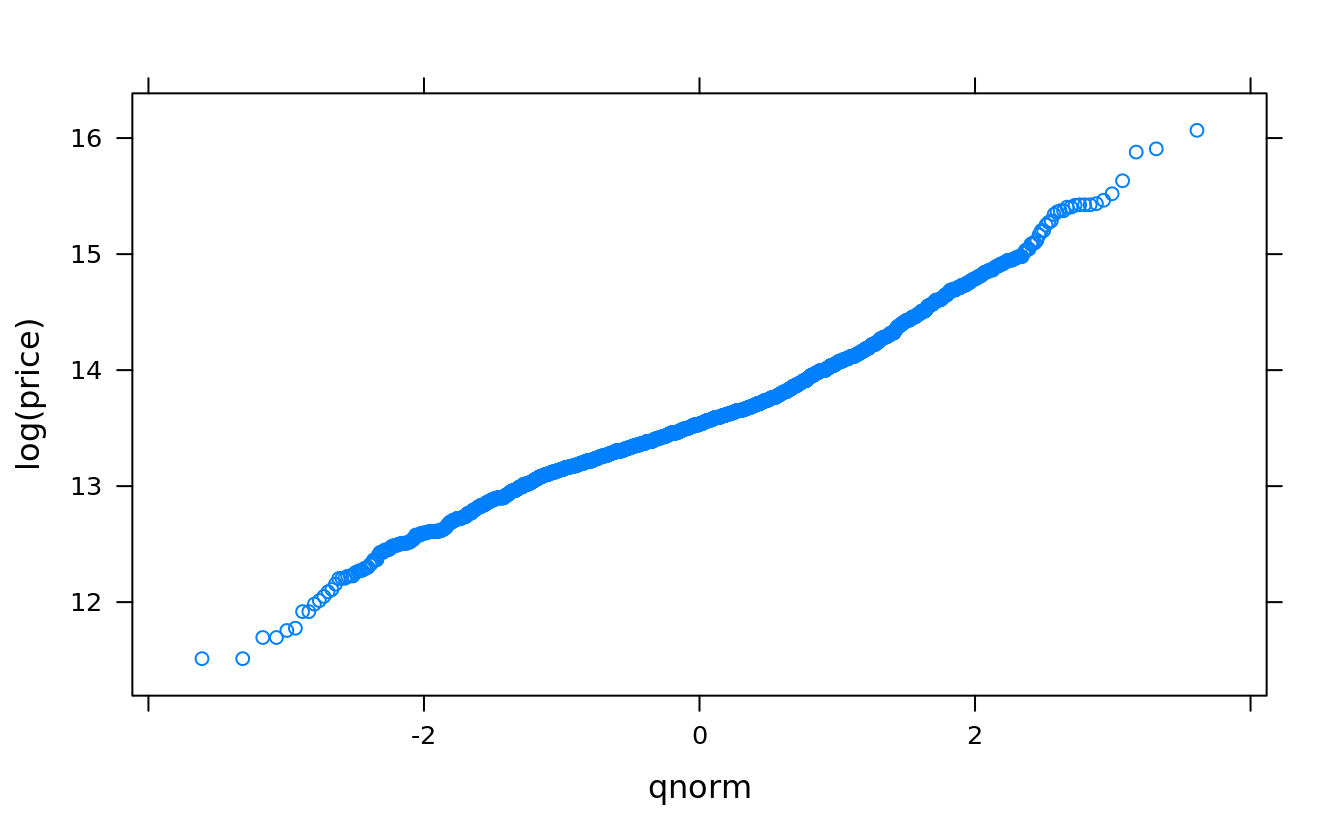
qqmath(~log(price), groups = bedrooms, auto.key = T, type = "smooth",
subset(san, !is.na(bedrooms) & bedrooms > 0 & bedrooms < 7))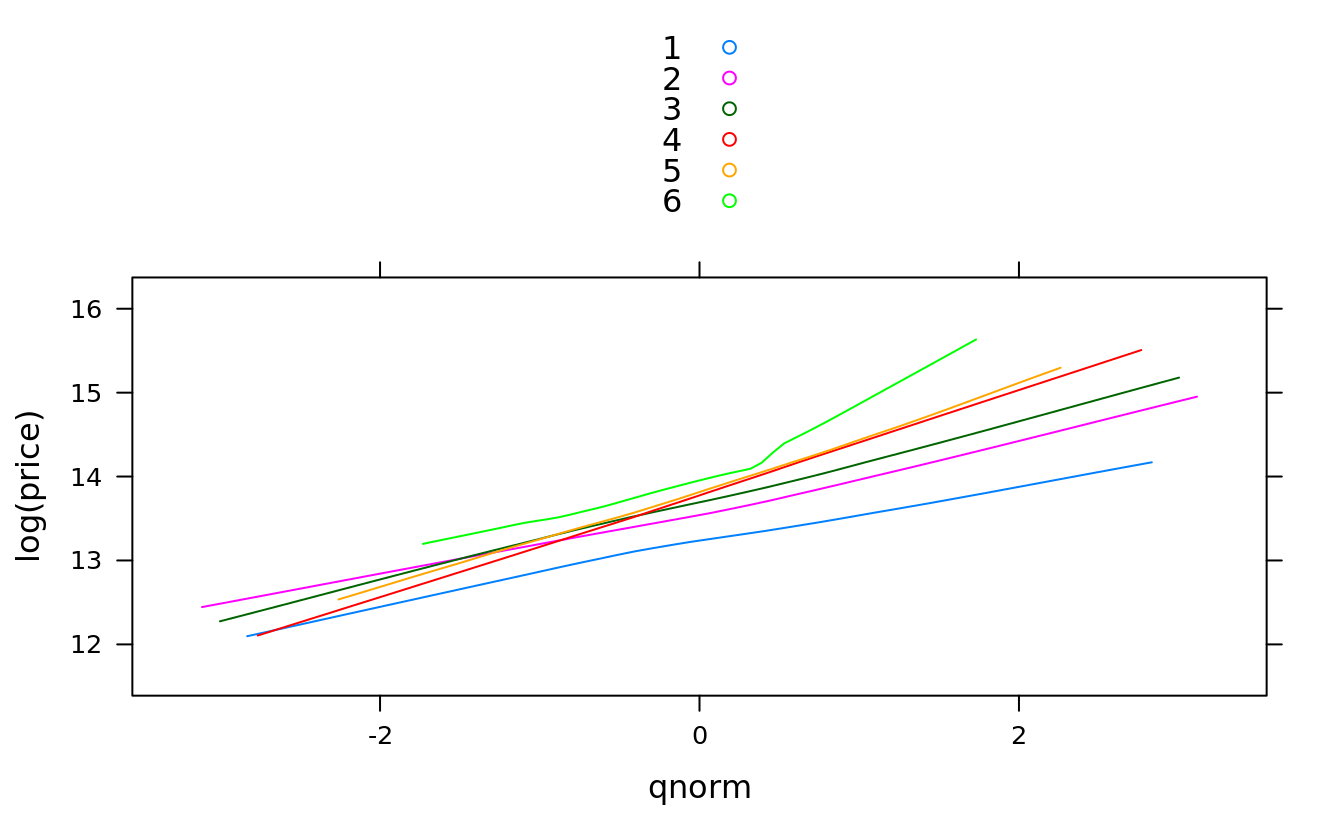
#using subset parameter causes unused factor levels:
qqmath(~log(price), groups = bedrooms, san, auto.key = T, type = "smooth",
subset = !is.na(bedrooms) & bedrooms > 0 & bedrooms < 7)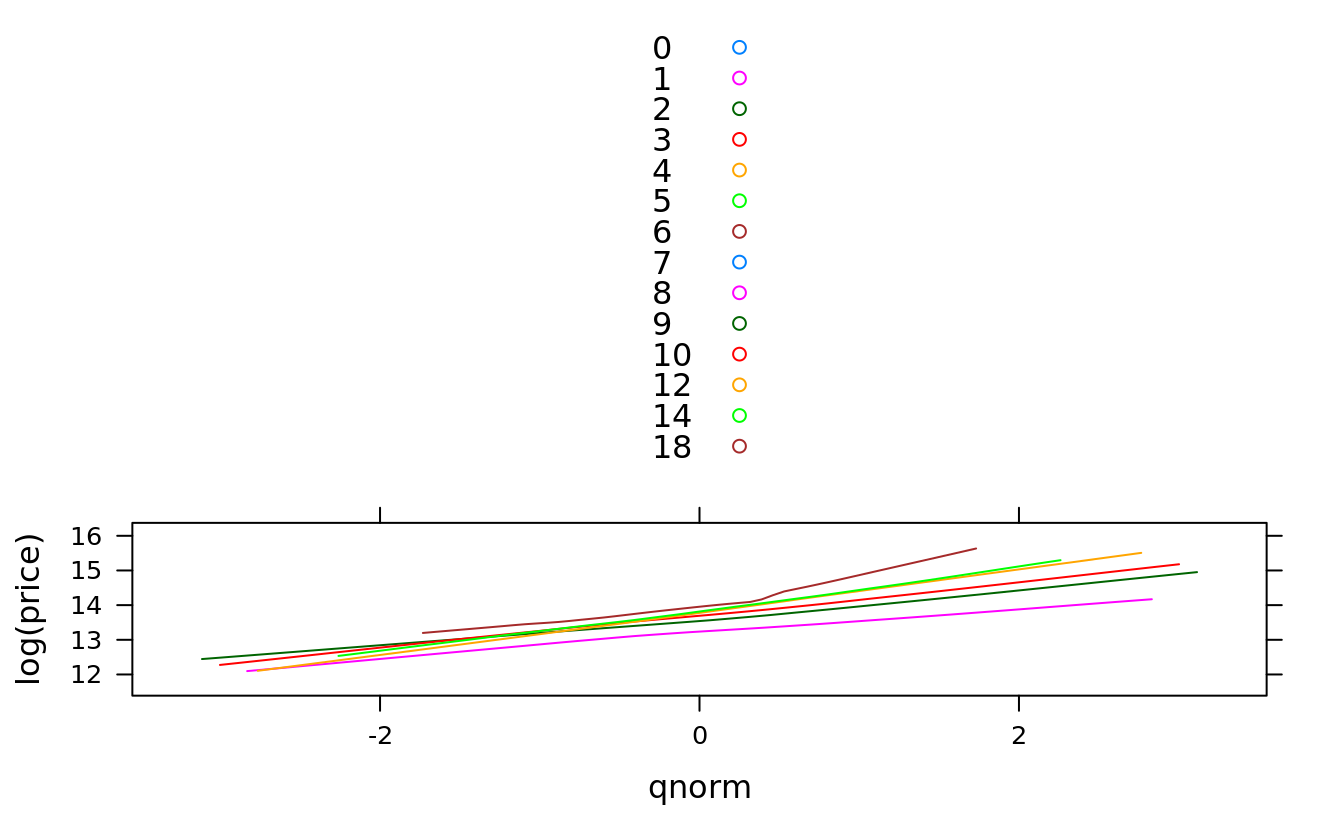
#we use subset parameter because NA is cleaned up:
qqmath(~log(price), groups = Hmisc::cut2(squarefeet, g = 6), san,
subset = !is.na(squarefeet), type = "smooth", auto.key = T)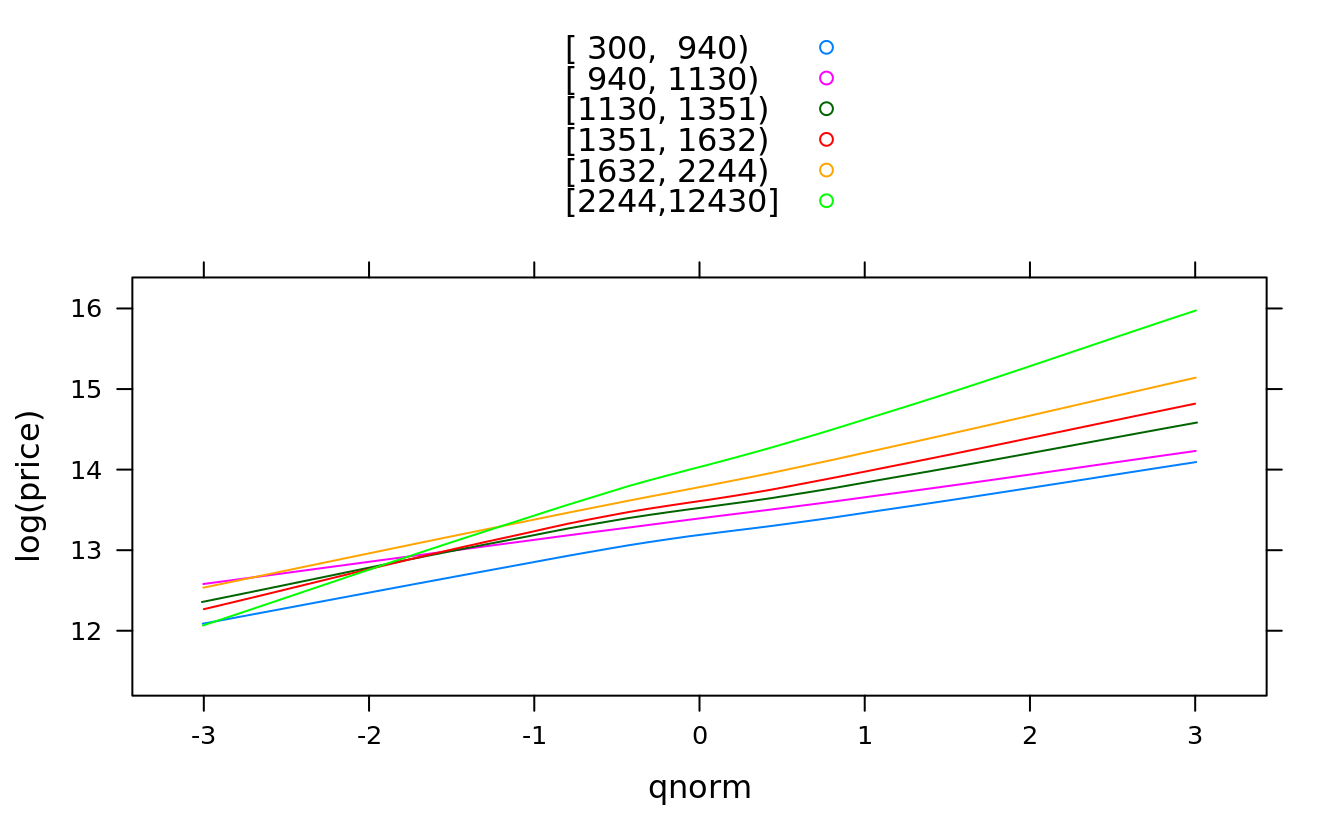
5.10 lattice: bivariate plot
5.10.1 scatter plot
xyplot(Turnout ~ Income, obama_vs_mccain, col = "violet", pch = 20)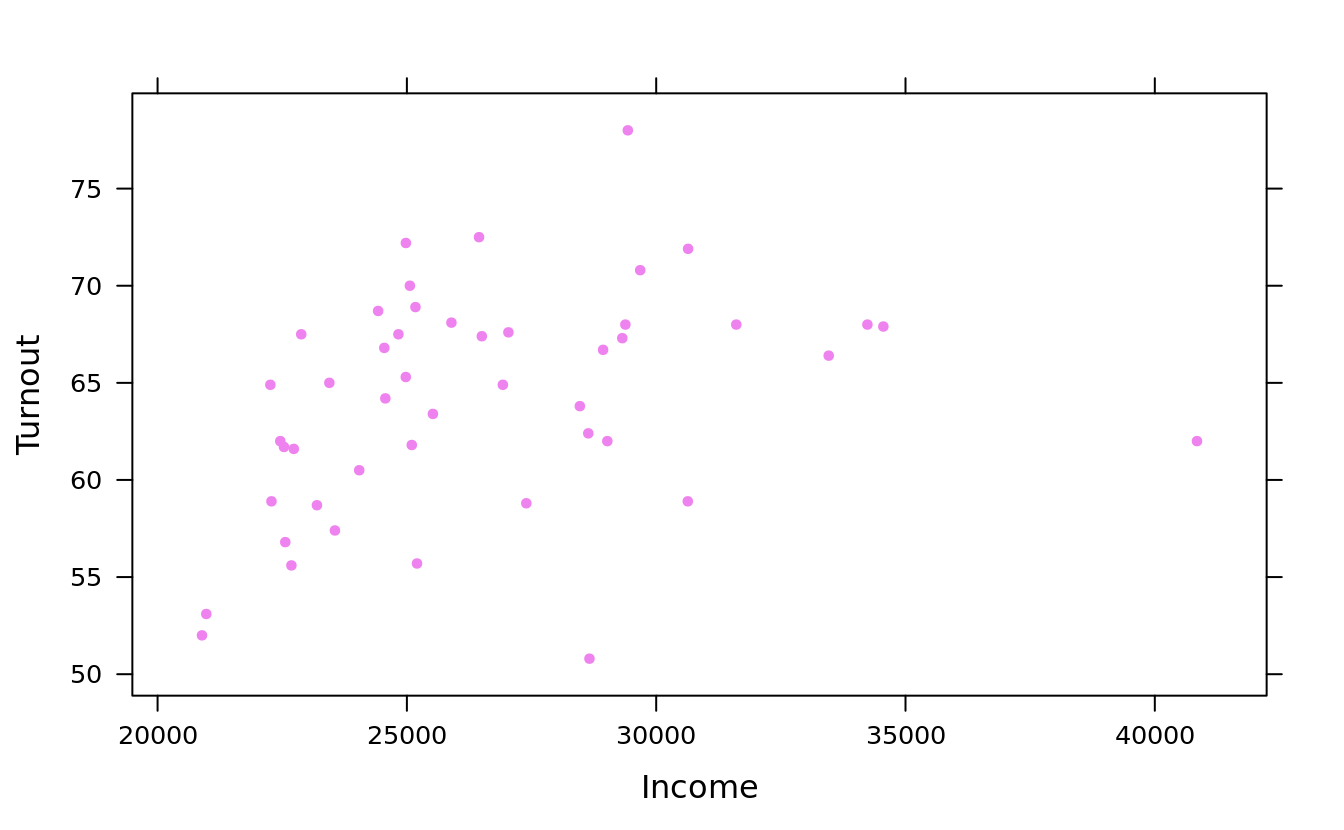
## use `list(log = TRUE)` to log-scale both axes
xyplot(Turnout ~ Income, obama_vs_mccain, scales = list(y = list(log = TRUE))) 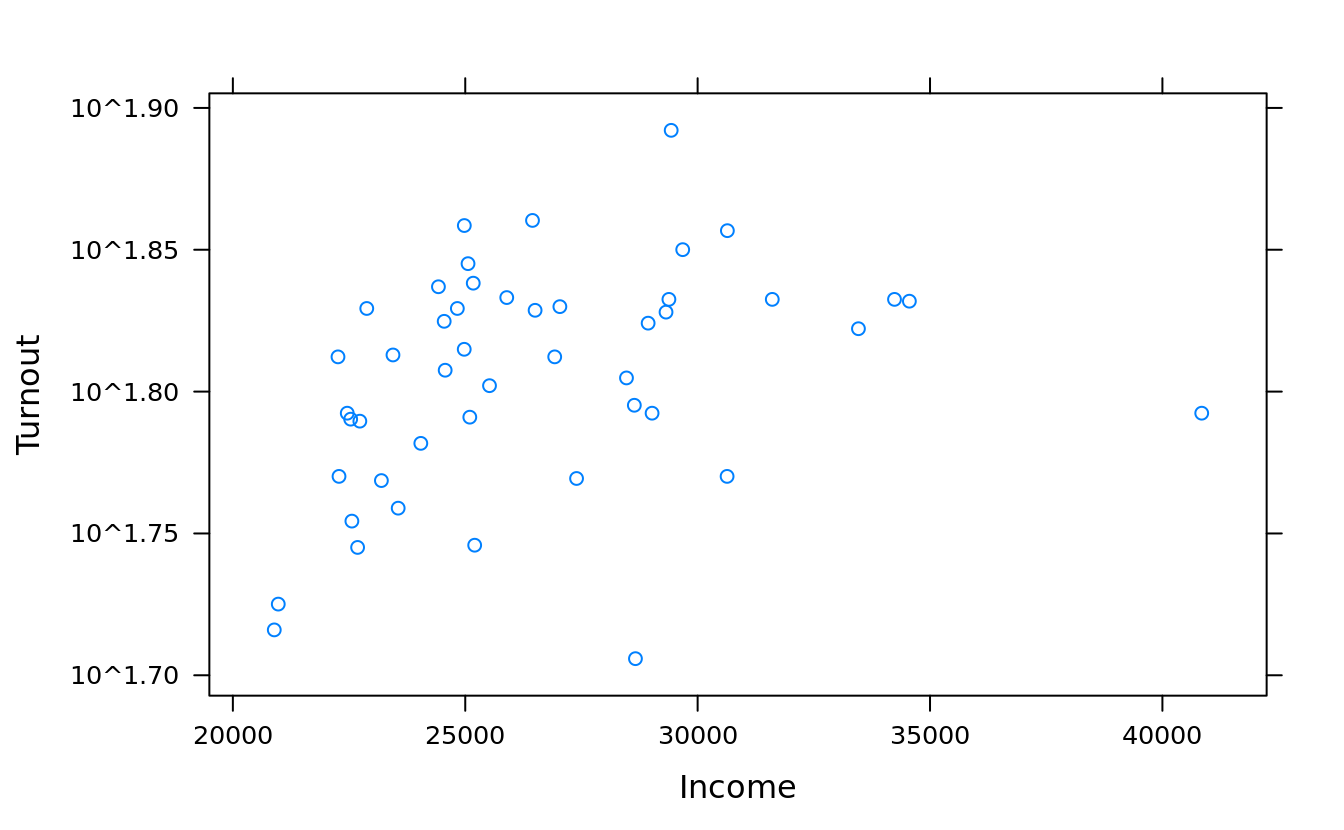
#relation = "same" means that each panel shares the same axes.
#alternating = T(default) means axis ticks for each panel are drawn on alternating sides of the plot, otherwise just the left and bottom
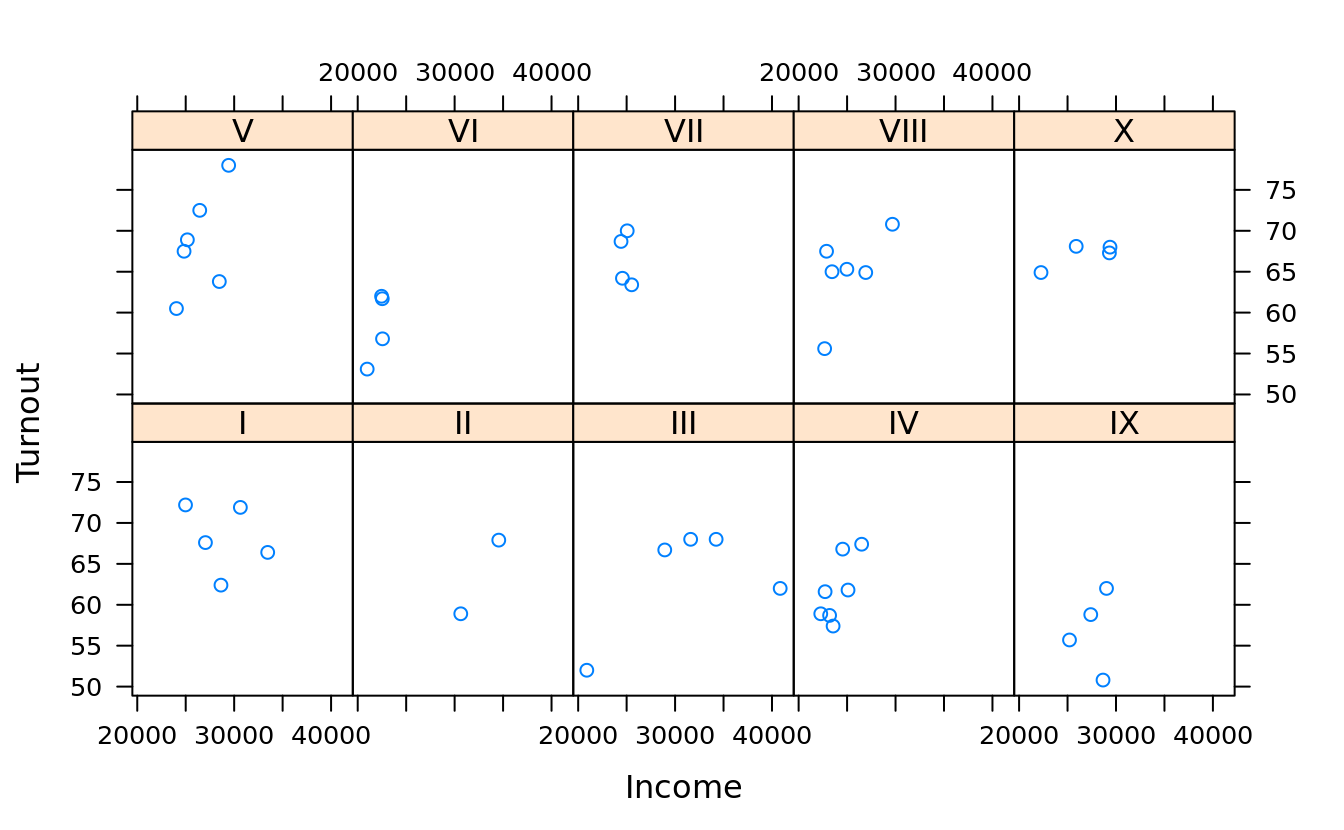
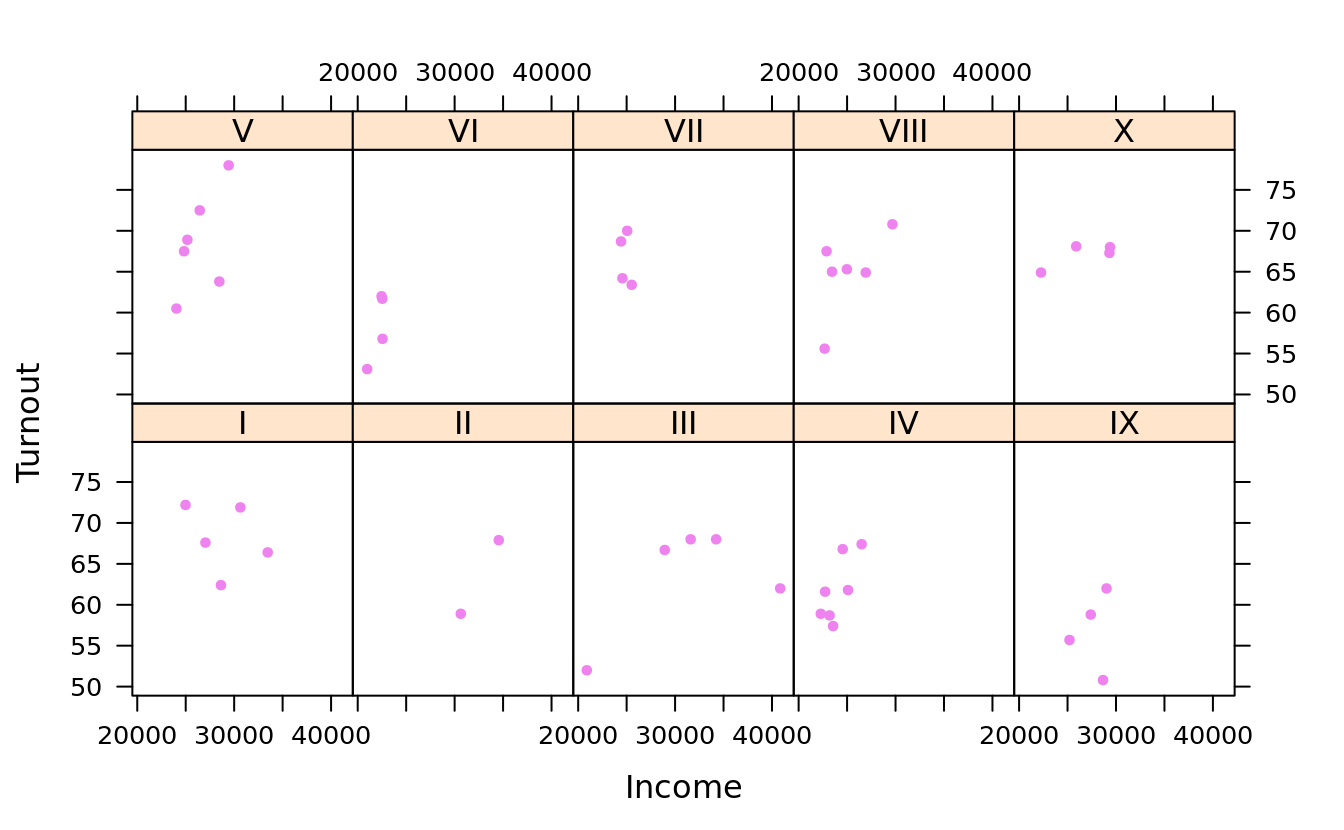
xyplot(Turnout ~ Income | Region, obama_vs_mccain, layout = c(5, 2),
scales = list(log = T, relation = "same", alternating = F))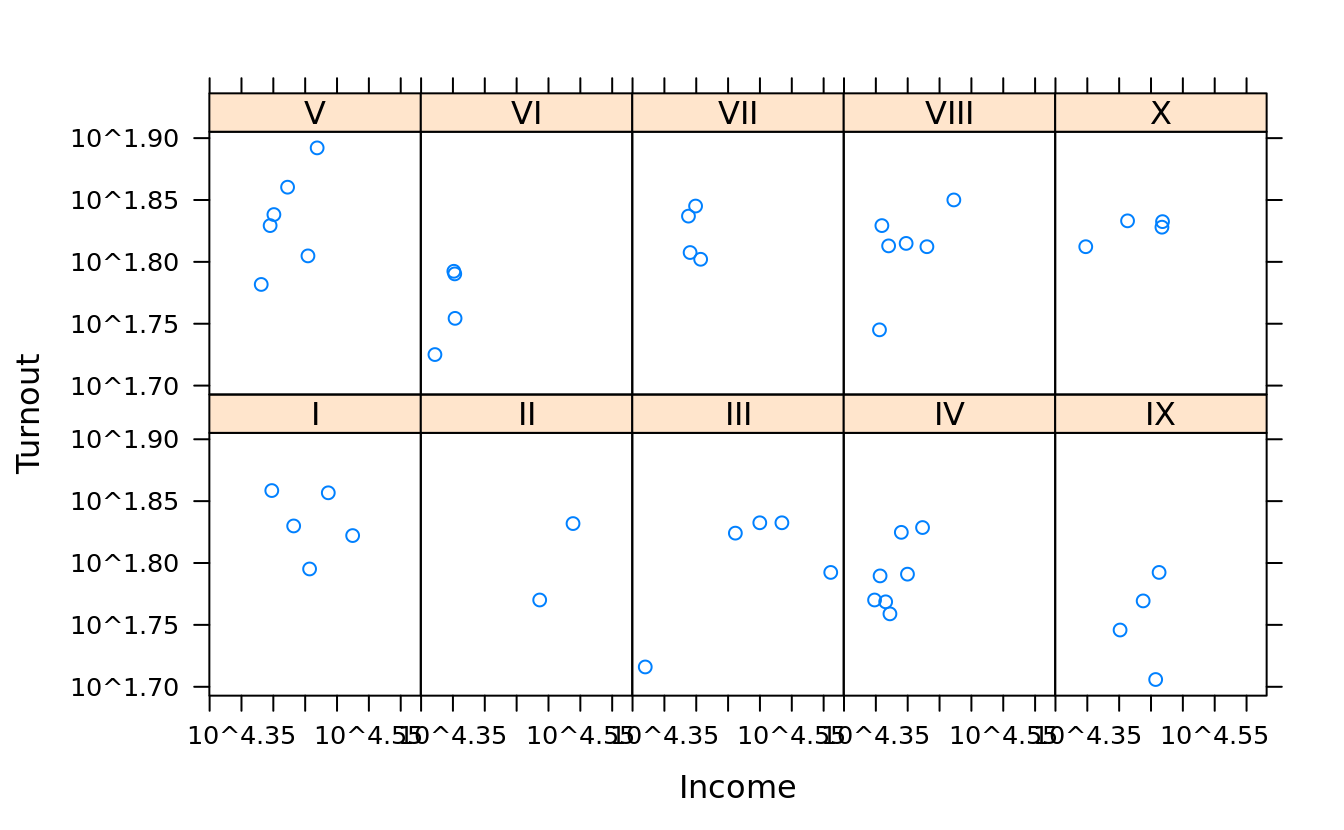
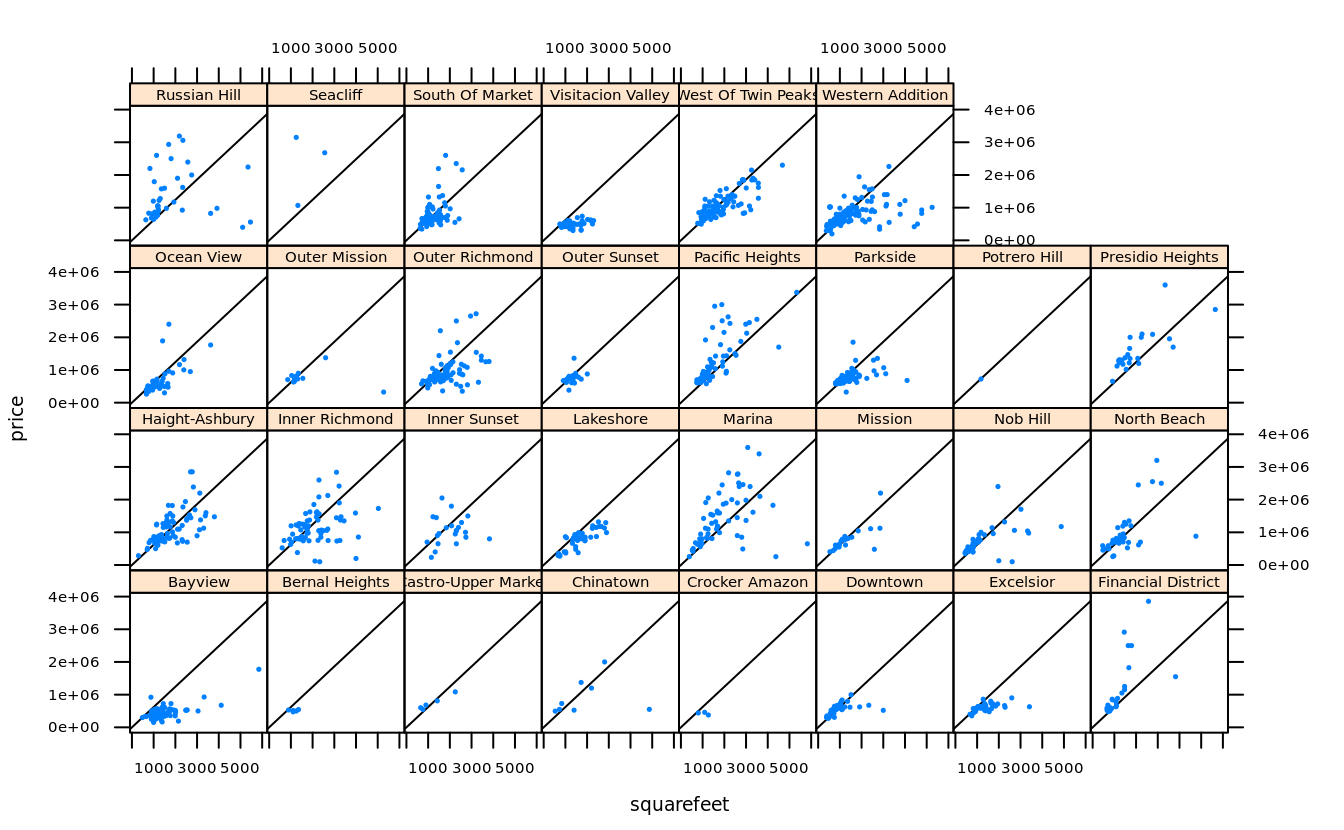
dollars.per.squarefoot <- with(san, mean(price / squarefeet, na.rm = T))
zips = subset(san, !is.na(squarefeet), zip) %>% table() %>% sort() %>% head(4*5) %>% names() %>% as.integer()
san.subset <- subset(san, zip %in% zips & price < 4000000 & squarefeet < 6000)
trellis.par.set(fontsize = list(text = 7))
xyplot(price~squarefeet | neighborhood, san.subset, pch = 19, cex = .2,
strip = strip.custom(strip.levels = T, horizontal = T,
par.strip.text = list(cex = .8)),
panel = function(...) {
panel.abline(a = 0, b = dollars.per.squarefoot)
panel.xyplot(...)
}
)
5.10.3 box plot
“bw” is short for “b (box) and w (whisker)”,
table(cut(san$date,"month"))
#>
#> 2008-02-01 2008-03-01 2008-04-01 2008-05-01 2008-06-01 2008-07-01 2008-08-01
#> 139 230 267 253 237 198 253
#> 2008-09-01 2008-10-01 2008-11-01 2008-12-01 2009-01-01 2009-02-01 2009-03-01
#> 223 272 118 181 114 123 142
#> 2009-04-01 2009-05-01 2009-06-01 2009-07-01
#> 116 180 150 85
bwplot(log(price) ~ cut(date,"month"), san, scales = list(x = list(rot = 90)))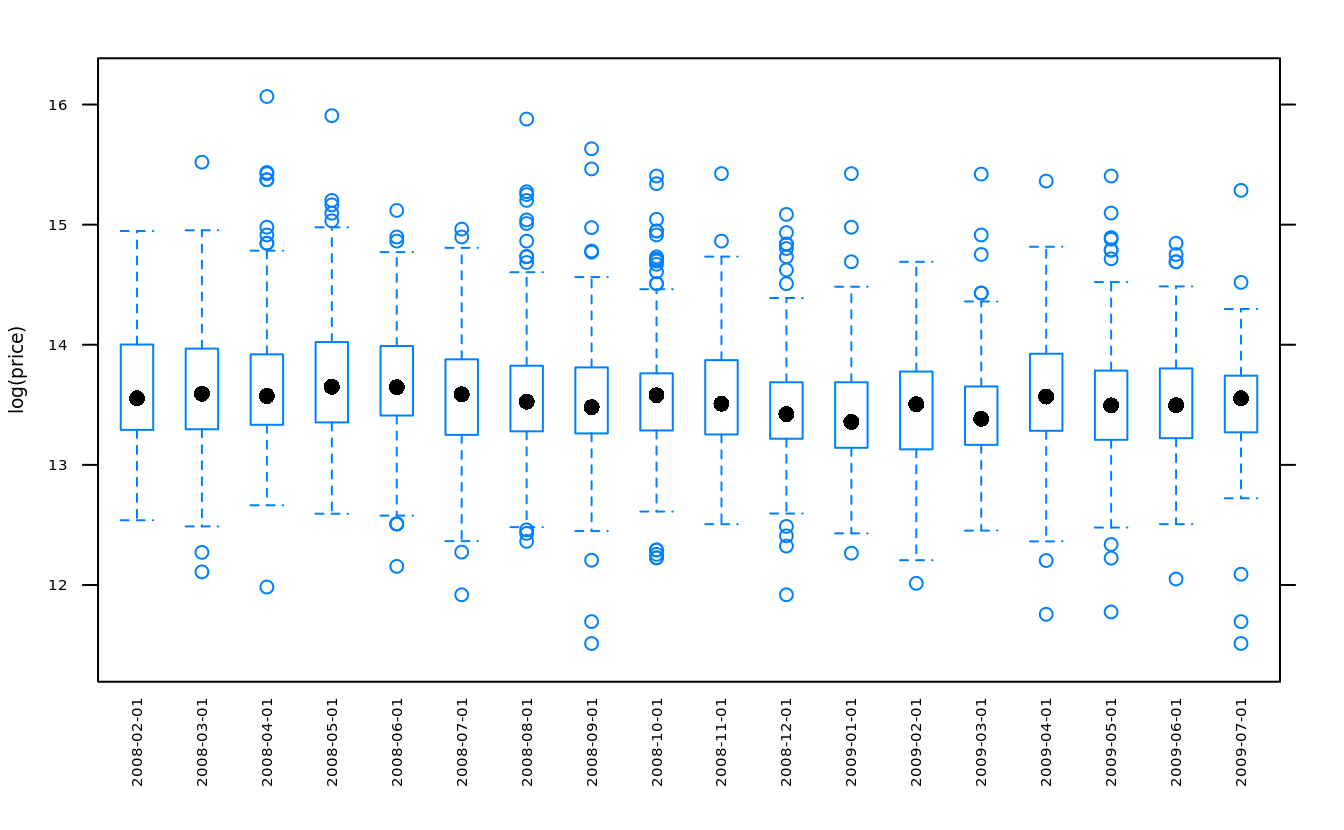
5.11 lattice: others
5.11.1 trivariate plot
levelplot(price~latitude + longitude, san)
levelplot(with(san, table(cut(longitude,40), cut(latitude,40))),
scales = list(x = list(rot = 90, cex = .5), y = list(cex = .5)))
levelplot(
with(san, tapply(price,list(cut(longitude, 40), cut(latitude, 40)), mean)),
scales = list(draw = F))
levelplot(with(san, table(cut(longitude, 25),
cut(latitude, 25),
ifelse(bedrooms < 5, bedrooms, 5))),
scales = list(draw = F))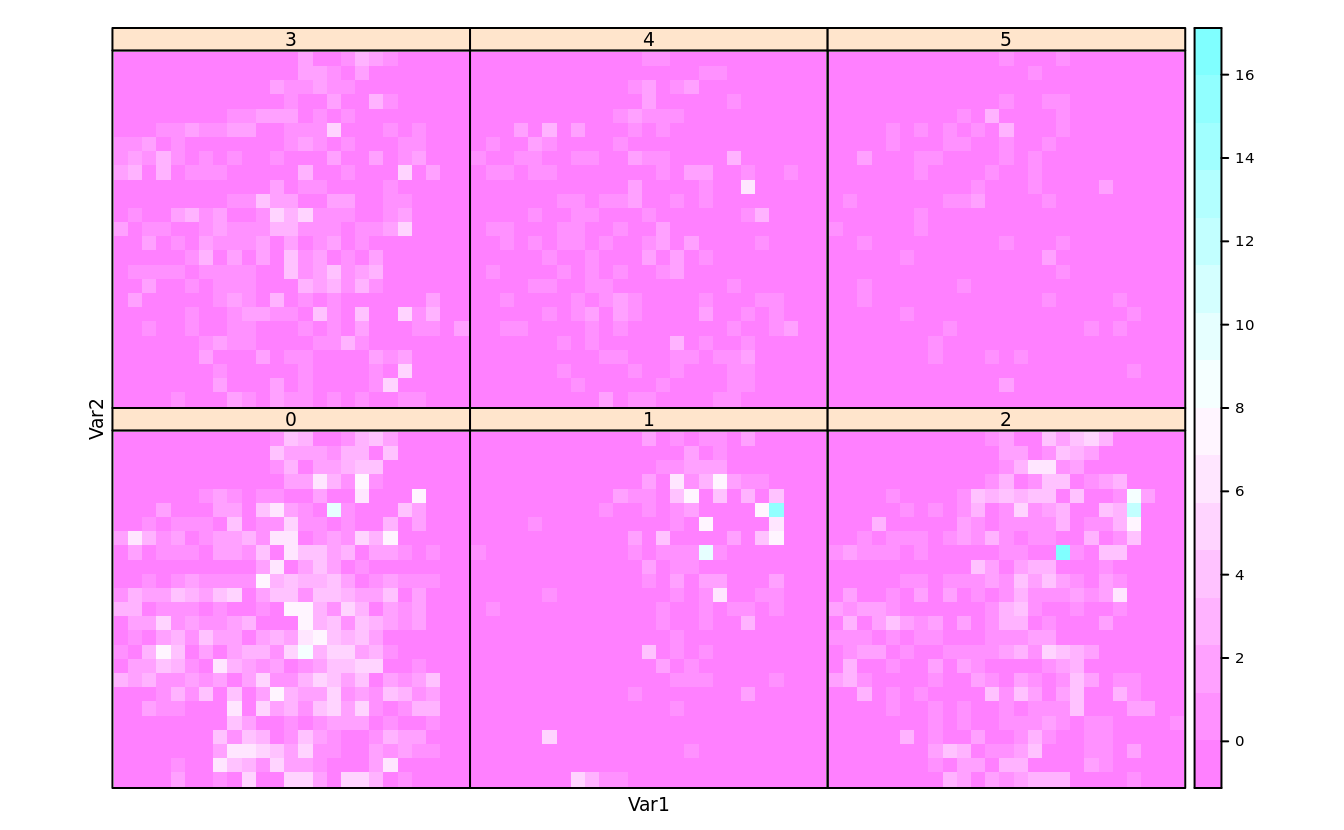
lattice::contourplot()lattice::cloud()lattice::wireframe()
5.11.2 residual and fit-spread (RFS) plot
model = lm(runs~singles + doubles + triples + stolenbases + homeruns + walks
+ hitbypitch + sacrificeflies + caughtstealing, team.batting.00to08)rfs(model)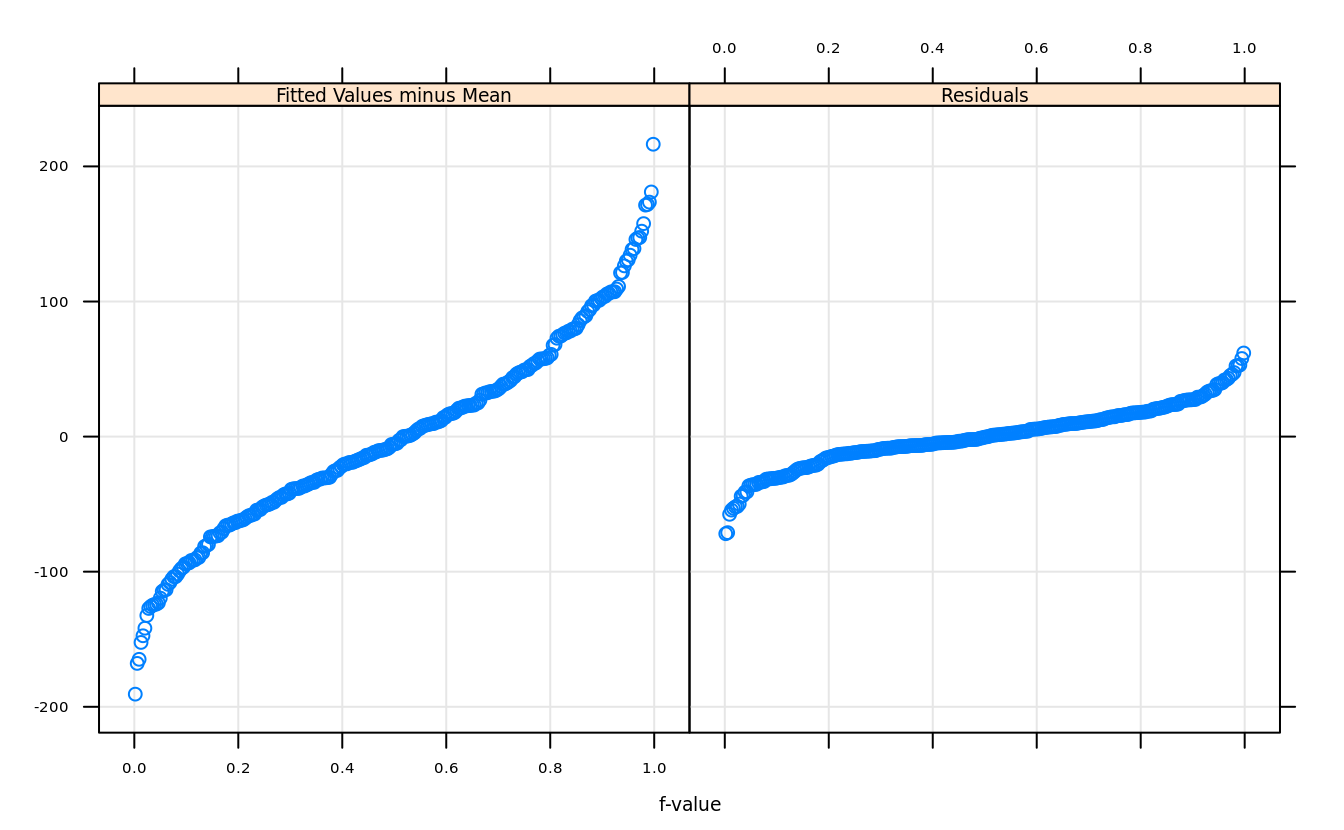
rfs(model,distribution = qnorm)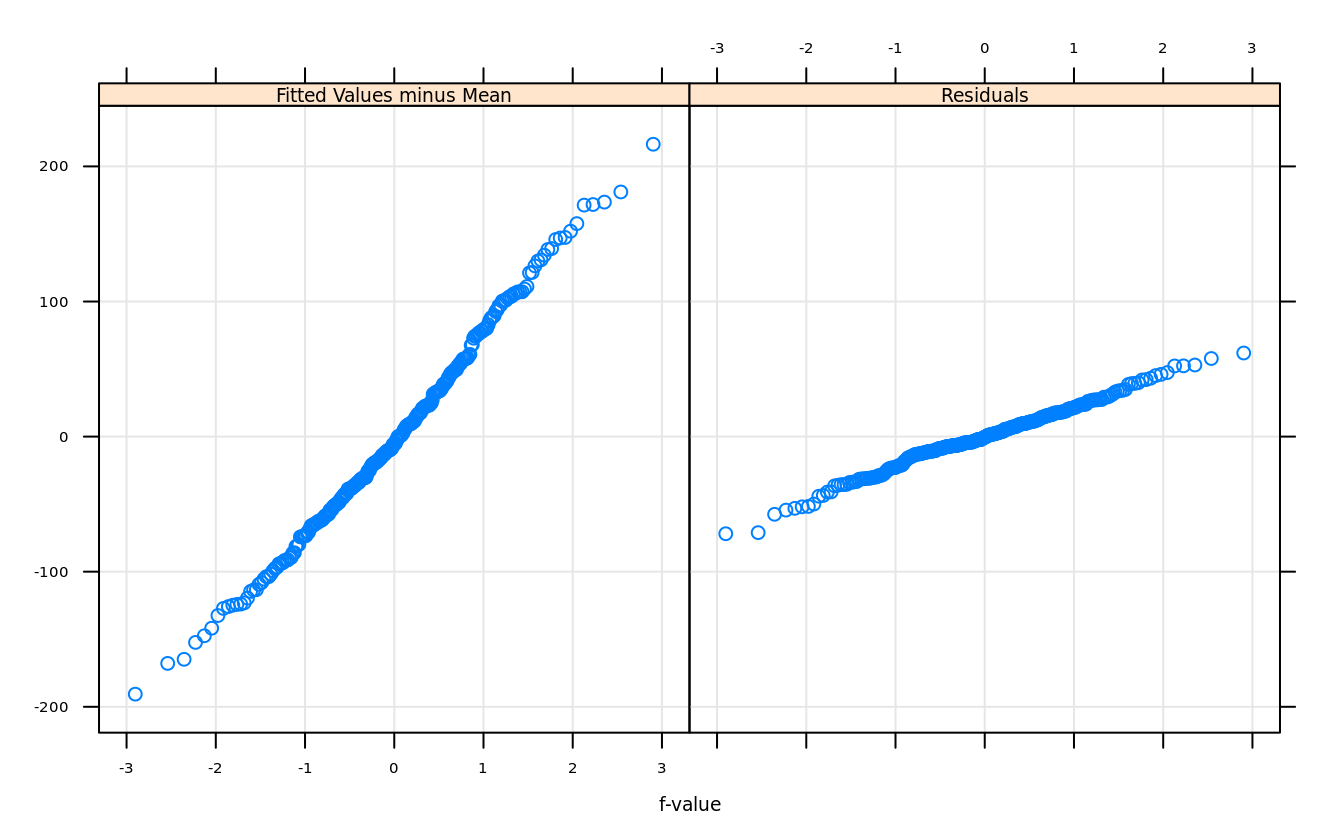
5.11.3 get parameters
trellis.par.get("clip")
#> $panel
#> [1] "on"
#>
#> $strip
#> [1] "on"
head(trellis.par.get(), 3)
#> $grid.pars
#> list()
#>
#> $fontsize
#> $fontsize$text
#> [1] 7
#>
#> $fontsize$points
#> [1] 8
#>
#>
#> $background
#> $background$alpha
#> [1] 1
#>
#> $background$col
#> [1] "transparent"5.11.6 deep diving
There are a total of 378 parameters. However, there are only 46 unique parameters within these groups:
length(unlist(trellis.par.get()))
#> [1] 382
(function() {
n = names(trellis.par.get())
p = NA
for (i in 1:34) p = c(p,names(trellis.par.get(n[i])))
length(table(p))
})()
#> [1] 48there are two GitHub repo, the former releases to CRAN↩︎