3.3 Attributes
You might have noticed that the set of atomic vectors does not include a number of important data structures like matrices, arrays, factors, or date-times. These types are built on top of atomic vectors by adding attributes. In this section, you’ll learn the basics of attributes, and how the dim attribute makes matrices and arrays. In the next section you’ll learn how the class attribute is used to create S3 vectors, including factors, dates, and date-times.
3.3.1 Getting and setting
You can think of attributes as name-value pairs16 that attach metadata to an object. Individual attributes can be retrieved and modified with attr(), or retrieved en masse with attributes(), and set en masse with structure().
a <- 1:3
attr(a, "x") <- "abcdef"
attr(a, "x")
#> [1] "abcdef"
attr(a, "y") <- 4:6
str(attributes(a))
#> List of 2
#> $ x: chr "abcdef"
#> $ y: int [1:3] 4 5 6
# Or equivalently
a <- structure(
1:3,
x = "abcdef",
y = 4:6
)
str(attributes(a))
#> List of 2
#> $ x: chr "abcdef"
#> $ y: int [1:3] 4 5 6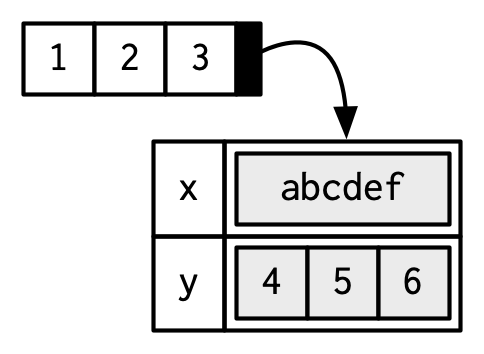
Attributes should generally be thought of as ephemeral. For example, most attributes are lost by most operations:
attributes(a[1])
#> NULL
attributes(sum(a))
#> NULLThere are only two attributes that are routinely preserved:
- names, a character vector giving each element a name.
- dim, short for dimensions, an integer vector, used to turn vectors into matrices or arrays.
To preserve other attributes, you’ll need to create your own S3 class, the topic of Chapter 13.
3.3.2 Names
You can name a vector in three ways:
# When creating it:
x <- c(a = 1, b = 2, c = 3)
# By assigning a character vector to names()
x <- 1:3
names(x) <- c("a", "b", "c")
# Inline, with setNames():
x <- setNames(1:3, c("a", "b", "c"))Avoid using attr(x, "names") as it requires more typing and is less readable than names(x). You can remove names from a vector by using unname(x) or names(x) <- NULL.
To be technically correct, when drawing the named vector x, I should draw it like so:
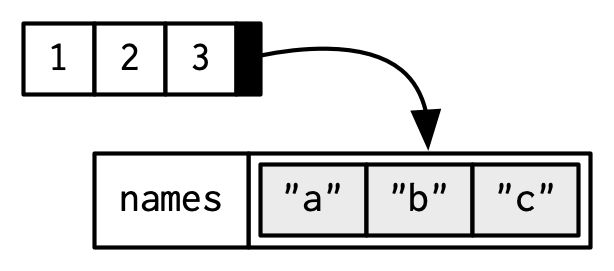
However, names are so special and so important, that unless I’m trying specifically to draw attention to the attributes data structure, I’ll use them to label the vector directly:
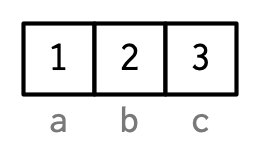
To be useful with character subsetting (e.g. Section 4.5.1) names should be unique, and non-missing, but this is not enforced by R. Depending on how the names are set, missing names may be either "" or NA_character_. If all names are missing, names() will return NULL.
3.3.3 Dimensions
Adding a dim attribute to a vector allows it to behave like a 2-dimensional matrix or a multi-dimensional array. Matrices and arrays are primarily mathematical and statistical tools, not programming tools, so they’ll be used infrequently and only covered briefly in this book. Their most important feature is multidimensional subsetting, which is covered in Section 4.2.3.
You can create matrices and arrays with matrix() and array(), or by using the assignment form of dim():
# Two scalar arguments specify row and column sizes
a <- matrix(1:6, nrow = 2, ncol = 3)
a
#> [,1] [,2] [,3]
#> [1,] 1 3 5
#> [2,] 2 4 6
# One vector argument to describe all dimensions
b <- array(1:12, c(2, 3, 2))
b
#> , , 1
#>
#> [,1] [,2] [,3]
#> [1,] 1 3 5
#> [2,] 2 4 6
#>
#> , , 2
#>
#> [,1] [,2] [,3]
#> [1,] 7 9 11
#> [2,] 8 10 12
# You can also modify an object in place by setting dim()
c <- 1:6
dim(c) <- c(3, 2)
c
#> [,1] [,2]
#> [1,] 1 4
#> [2,] 2 5
#> [3,] 3 6Many of the functions for working with vectors have generalisations for matrices and arrays:
| Vector | Matrix | Array |
|---|---|---|
names() |
rownames(), colnames() |
dimnames() |
length() |
nrow(), ncol() |
dim() |
c() |
rbind(), cbind() |
abind::abind() |
| — | t() |
aperm() |
is.null(dim(x)) |
is.matrix() |
is.array() |
A vector without a dim attribute set is often thought of as 1-dimensional, but actually has NULL dimensions. You also can have matrices with a single row or single column, or arrays with a single dimension. They may print similarly, but will behave differently. The differences aren’t too important, but it’s useful to know they exist in case you get strange output from a function (tapply() is a frequent offender). As always, use str() to reveal the differences.
str(1:3) # 1d vector
#> int [1:3] 1 2 3
str(matrix(1:3, ncol = 1)) # column vector
#> int [1:3, 1] 1 2 3
str(matrix(1:3, nrow = 1)) # row vector
#> int [1, 1:3] 1 2 3
str(array(1:3, 3)) # "array" vector
#> int [1:3(1d)] 1 2 33.3.4 Exercises
How is
setNames()implemented? How isunname()implemented? Read the source code.What does
dim()return when applied to a 1-dimensional vector? When might you useNROW()orNCOL()?How would you describe the following three objects? What makes them different from
1:5?x1 <- array(1:5, c(1, 1, 5)) x2 <- array(1:5, c(1, 5, 1)) x3 <- array(1:5, c(5, 1, 1))An early draft used this code to illustrate
structure():structure(1:5, comment = "my attribute") #> [1] 1 2 3 4 5But when you print that object you don’t see the comment attribute. Why? Is the attribute missing, or is there something else special about it? (Hint: try using help.)