9.2 My first functional: map()
The most fundamental functional is purrr::map()38. It takes a vector and a function, calls the function once for each element of the vector, and returns the results in a list. In other words, map(1:3, f) is equivalent to list(f(1), f(2), f(3)).
triple <- function(x) x * 3
map(1:3, triple)
#> [[1]]
#> [1] 3
#>
#> [[2]]
#> [1] 6
#>
#> [[3]]
#> [1] 9Or, graphically:
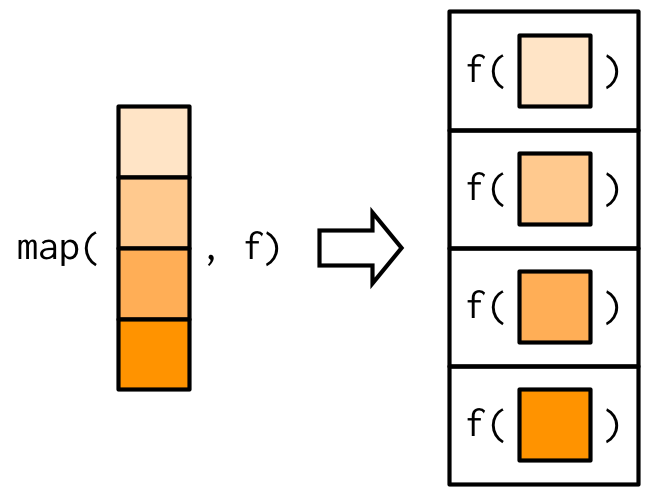
The implementation of map() is quite simple. We allocate a list the same length as the input, and then fill in the list with a for loop. The heart of the implementation is only a handful of lines of code:
simple_map <- function(x, f, ...) {
out <- vector("list", length(x))
for (i in seq_along(x)) {
out[[i]] <- f(x[[i]], ...)
}
out
}The real purrr::map() function has a few differences: it is written in C to eke out every last iota of performance, preserves names, and supports a few shortcuts that you’ll learn about in Section 9.2.2.
The base equivalent to map() is lapply(). The only difference is that lapply() does not support the helpers that you’ll learn about below, so if you’re only using map() from purrr, you can skip the additional dependency and use lapply() directly.
9.2.1 Producing atomic vectors
map() returns a list, which makes it the most general of the map family because you can put anything in a list. But it is inconvenient to return a list when a simpler data structure would do, so there are four more specific variants: map_lgl(), map_int(), map_dbl(), and map_chr(). Each returns an atomic vector of the specified type:
# map_chr() always returns a character vector
map_chr(mtcars, typeof)
#> mpg cyl disp hp drat wt qsec vs
#> "double" "double" "double" "double" "double" "double" "double" "double"
#> am gear carb
#> "double" "double" "double"
# map_lgl() always returns a logical vector
map_lgl(mtcars, is.double)
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
# map_int() always returns a integer vector
n_unique <- function(x) length(unique(x))
map_int(mtcars, n_unique)
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> 25 3 27 22 22 29 30 2 2 3 6
# map_dbl() always returns a double vector
map_dbl(mtcars, mean)
#> mpg cyl disp hp drat wt qsec vs am gear
#> 20.091 6.188 230.722 146.688 3.597 3.217 17.849 0.438 0.406 3.688
#> carb
#> 2.812purrr uses the convention that suffixes, like _dbl(), refer to the output. All map_*() functions can take any type of vector as input. These examples rely on two facts: mtcars is a data frame, and data frames are lists containing vectors of the same length. This is more obvious if we draw a data frame with the same orientation as vector:
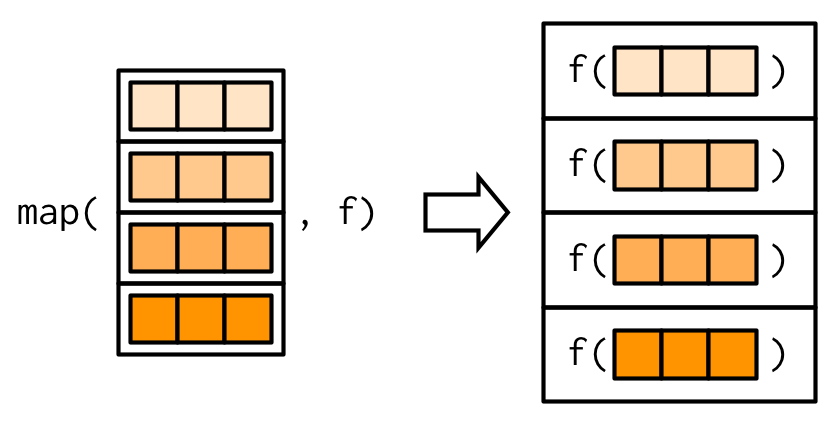
All map functions always return an output vector the same length as the input, which implies that each call to .f must return a single value. If it does not, you’ll get an error:
pair <- function(x) c(x, x)
map_dbl(1:2, pair)
#> Error: Result 1 must be a single double, not an integer vector of length 2This is similar to the error you’ll get if .f returns the wrong type of result:
map_dbl(1:2, as.character)
#> Error: Can't coerce element 1 from a character to a doubleIn either case, it’s often useful to switch back to map(), because map() can accept any type of output. That allows you to see the problematic output, and figure out what to do with it.
map(1:2, pair)
#> [[1]]
#> [1] 1 1
#>
#> [[2]]
#> [1] 2 2
map(1:2, as.character)
#> [[1]]
#> [1] "1"
#>
#> [[2]]
#> [1] "2"Base R has two apply functions that can return atomic vectors: sapply() and vapply(). I recommend that you avoid sapply() because it tries to simplify the result, so it can return a list, a vector, or a matrix. This makes it difficult to program with, and it should be avoided in non-interactive settings. vapply() is safer because it allows you to provide a template, FUN.VALUE, that describes the output shape. If you don’t want to use purrr, I recommend you always use vapply() in your functions, not sapply(). The primary downside of vapply() is its verbosity: for example, the equivalent to map_dbl(x, mean, na.rm = TRUE) is vapply(x, mean, na.rm = TRUE, FUN.VALUE = double(1)).
9.2.2 Anonymous functions and shortcuts
Instead of using map() with an existing function, you can create an inline anonymous function (as mentioned in Section 6.2.3):
map_dbl(mtcars, function(x) length(unique(x)))
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> 25 3 27 22 22 29 30 2 2 3 6Anonymous functions are very useful, but the syntax is verbose. So purrr supports a special shortcut:
map_dbl(mtcars, ~ length(unique(.x)))
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> 25 3 27 22 22 29 30 2 2 3 6This works because all purrr functions translate formulas, created by ~ (pronounced “twiddle”), into functions. You can see what’s happening behind the scenes by calling as_mapper():
as_mapper(~ length(unique(.x)))
#> <lambda>
#> function (..., .x = ..1, .y = ..2, . = ..1)
#> length(unique(.x))
#> attr(,"class")
#> [1] "rlang_lambda_function" "function"The function arguments look a little quirky but allow you to refer to . for one argument functions, .x and .y. for two argument functions, and ..1, ..2, ..3, etc, for functions with an arbitrary number of arguments. . remains for backward compatibility but I don’t recommend using it because it’s easily confused with the . used by magrittr’s pipe.
This shortcut is particularly useful for generating random data:
x <- map(1:3, ~ runif(2))
str(x)
#> List of 3
#> $ : num [1:2] 0.281 0.53
#> $ : num [1:2] 0.433 0.917
#> $ : num [1:2] 0.0275 0.8249Reserve this syntax for short and simple functions. A good rule of thumb is that if your function spans lines or uses {}, it’s time to give it a name.
The map functions also have shortcuts for extracting elements from a vector, powered by purrr::pluck(). You can use a character vector to select elements by name, an integer vector to select by position, or a list to select by both name and position. These are very useful for working with deeply nested lists, which often arise when working with JSON.
x <- list(
list(-1, x = 1, y = c(2), z = "a"),
list(-2, x = 4, y = c(5, 6), z = "b"),
list(-3, x = 8, y = c(9, 10, 11))
)
# Select by name
map_dbl(x, "x")
#> [1] 1 4 8
# Or by position
map_dbl(x, 1)
#> [1] -1 -2 -3
# Or by both
map_dbl(x, list("y", 1))
#> [1] 2 5 9
# You'll get an error if a component doesn't exist:
map_chr(x, "z")
#> Error: Result 3 must be a single string, not NULL of length 0
# Unless you supply a .default value
map_chr(x, "z", .default = NA)
#> [1] "a" "b" NAIn base R functions, like lapply(), you can provide the name of the function as a string. This isn’t tremendously useful as lapply(x, "f") is almost always equivalent to lapply(x, f) and is more typing.
9.2.3 Passing arguments with ...
It’s often convenient to pass along additional arguments to the function that you’re calling. For example, you might want to pass na.rm = TRUE along to mean(). One way to do that is with an anonymous function:
x <- list(1:5, c(1:10, NA))
map_dbl(x, ~ mean(.x, na.rm = TRUE))
#> [1] 3.0 5.5But because the map functions pass ... along, there’s a simpler form available:
map_dbl(x, mean, na.rm = TRUE)
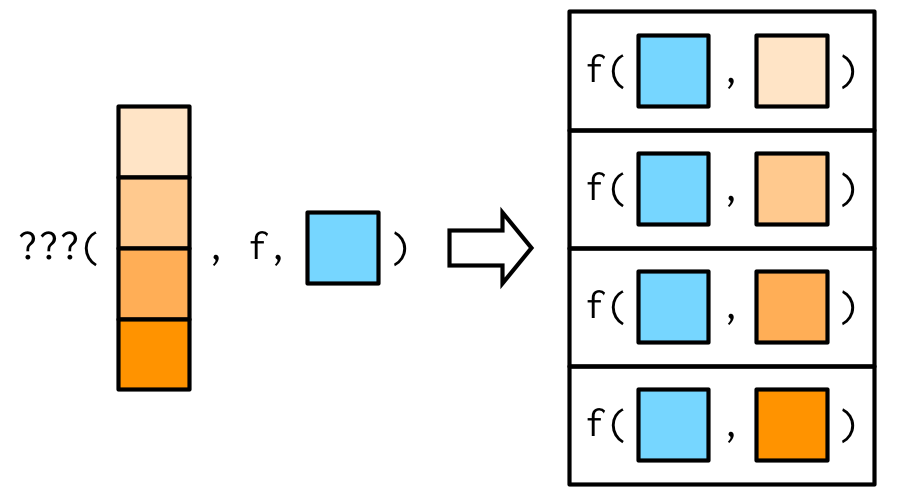
#> [1] 3.0 5.5This is easiest to understand with a picture: any arguments that come after f in the call to map() are inserted after the data in individual calls to f():
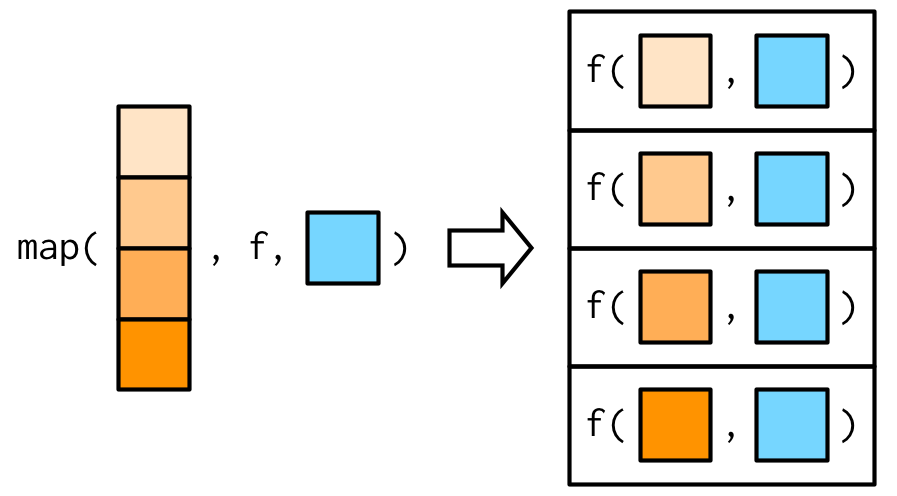
It’s important to note that these arguments are not decomposed; or said another way, map() is only vectorised over its first argument. If an argument after f is a vector, it will be passed along as is:
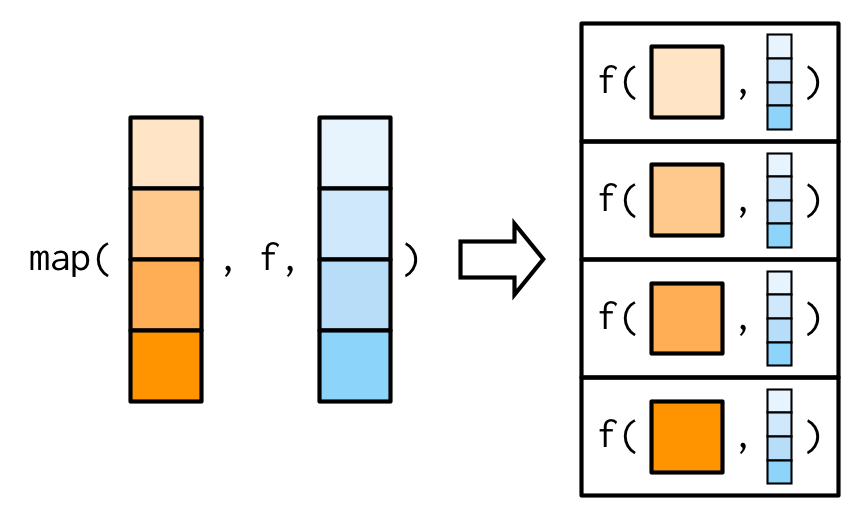
(You’ll learn about map variants that are vectorised over multiple arguments in Sections 9.4.2 and 9.4.5.)
Note there’s a subtle difference between placing extra arguments inside an anonymous function compared with passing them to map(). Putting them in an anonymous function means that they will be evaluated every time f() is executed, not just once when you call map(). This is easiest to see if we make the additional argument random:
plus <- function(x, y) x + y
x <- c(0, 0, 0, 0)
map_dbl(x, plus, runif(1))
#> [1] 0.0625 0.0625 0.0625 0.0625
map_dbl(x, ~ plus(.x, runif(1)))
#> [1] 0.903 0.132 0.629 0.9459.2.4 Argument names
In the diagrams, I’ve omitted argument names to focus on the overall structure. But I recommend writing out the full names in your code, as it makes it easier to read. map(x, mean, 0.1) is perfectly valid code, but will call mean(x[[1]], 0.1) so it relies on the reader remembering that the second argument to mean() is trim. To avoid unnecessary burden on the brain of the reader39, be kind and write map(x, mean, trim = 0.1).
This is the reason why the arguments to map() are a little odd: instead of being x and f, they are .x and .f. It’s easiest to see the problem that leads to these names using simple_map() defined above. simple_map() has arguments x and f so you’ll have problems whenever the function you are calling has arguments x or f:
boostrap_summary <- function(x, f) {
f(sample(x, replace = TRUE))
}
simple_map(mtcars, boostrap_summary, f = mean)
#> Error in mean.default(x[[i]], ...): 'trim' must be numeric of length oneThe error is a little bewildering until you remember that the call to simple_map() is equivalent to simple_map(x = mtcars, f = mean, bootstrap_summary) because named matching beats positional matching.
purrr functions reduce the likelihood of such a clash by using .f and .x instead of the more common f and x. Of course this technique isn’t perfect (because the function you are calling might still use .f and .x), but it avoids 99% of issues. The remaining 1% of the time, use an anonymous function.
Base functions that pass along ... use a variety of naming conventions to prevent undesired argument matching:
The apply family mostly uses capital letters (e.g.
XandFUN).transform()uses the more exotic prefix_: this makes the name non-syntactic so it must always be surrounded in`, as described in Section 2.2.1. This makes undesired matches extremely unlikely.Other functionals like
uniroot()andoptim()make no effort to avoid clashes but they tend to be used with specially created functions so clashes are less likely.
9.2.5 Varying another argument
So far the first argument to map() has always become the first argument to the function. But what happens if the first argument should be constant, and you want to vary a different argument? How do you get the result in this picture?
It turns out that there’s no way to do it directly, but there are two tricks you can use instead. To illustrate them, imagine I have a vector that contains a few unusual values, and I want to explore the effect of different amounts of trimming when computing the mean. In this case, the first argument to mean() will be constant, and I want to vary the second argument, trim.
trims <- c(0, 0.1, 0.2, 0.5)
x <- rcauchy(1000)The simplest technique is to use an anonymous function to rearrange the argument order:
map_dbl(trims, ~ mean(x, trim = .x)) #> [1] -0.3500 0.0434 0.0354 0.0502This is still a little confusing because I’m using both
xand.x. You can make it a little clearer by abandoning the~helper:map_dbl(trims, function(trim) mean(x, trim = trim)) #> [1] -0.3500 0.0434 0.0354 0.0502Sometimes, if you want to be (too) clever, you can take advantage of R’s flexible argument matching rules (as described in Section 6.8.2). For example, in this example you can rewrite
mean(x, trim = 0.1)asmean(0.1, x = x), so you could write the call tomap_dbl()as:map_dbl(trims, mean, x = x) #> [1] -0.3500 0.0434 0.0354 0.0502I don’t recommend this technique as it relies on the reader’s familiarity with both the argument order to
.f, and R’s argument matching rules.
You’ll see one more alternative in Section 9.4.5.
9.2.6 Exercises
Use
as_mapper()to explore how purrr generates anonymous functions for the integer, character, and list helpers. What helper allows you to extract attributes? Read the documentation to find out.map(1:3, ~ runif(2))is a useful pattern for generating random numbers, butmap(1:3, runif(2))is not. Why not? Can you explain why it returns the result that it does?Use the appropriate
map()function to:Compute the standard deviation of every column in a numeric data frame.
Compute the standard deviation of every numeric column in a mixed data frame. (Hint: you’ll need to do it in two steps.)
Compute the number of levels for every factor in a data frame.
The following code simulates the performance of a t-test for non-normal data. Extract the p-value from each test, then visualise.
trials <- map(1:100, ~ t.test(rpois(10, 10), rpois(7, 10)))The following code uses a map nested inside another map to apply a function to every element of a nested list. Why does it fail, and what do you need to do to make it work?
x <- list( list(1, c(3, 9)), list(c(3, 6), 7, c(4, 7, 6)) ) triple <- function(x) x * 3 map(x, map, .f = triple) #> Error in .f(.x[[i]], ...): unused argument (function (.x, .f, ...) #> { #> .f <- as_mapper(.f, ...) #> .Call(map_impl, environment(), ".x", ".f", "list") #> })Use
map()to fit linear models to themtcarsdataset using the formulas stored in this list:formulas <- list( mpg ~ disp, mpg ~ I(1 / disp), mpg ~ disp + wt, mpg ~ I(1 / disp) + wt )Fit the model
mpg ~ dispto each of the bootstrap replicates ofmtcarsin the list below, then extract the of the model fit (Hint: you can compute the withsummary().)bootstrap <- function(df) { df[sample(nrow(df), replace = TRUE), , drop = FALSE] } bootstraps <- map(1:10, ~ bootstrap(mtcars))