8.1 Laying out plots side by side
Often, one wants to show two or more plots side by side to show different aspects of the same story in a compelling way. This is the scenario that patchwork was build to solve. At it’s heart, patchwork is a package that extends ggplot2’s use of the + operator to work between multiple plots, as well as add additional operators for specialized compositions and working with compositions of plots.
As an example of the most basic use of patchwork, we’ll use the following 4 plots of the mpg dataset
p1 <- ggplot(mpg) +
geom_point(aes(x = displ, y = hwy))
p2 <- ggplot(mpg) +
geom_bar(aes(x = as.character(year), fill = drv), position = "dodge") +
labs(x = "year")
p3 <- ggplot(mpg) +
geom_density(aes(x = hwy, fill = drv), colour = NA) +
facet_grid(rows = vars(drv))
p4 <- ggplot(mpg) +
stat_summary(aes(x = drv, y = hwy, fill = drv), geom = "col", fun.data = mean_se) +
stat_summary(aes(x = drv, y = hwy), geom = "errorbar", fun.data = mean_se, width = 0.5)The most simple use of patchwork is to use + to add plots together thus creating an assemble of plots to display together:
library(patchwork)
p1 + p2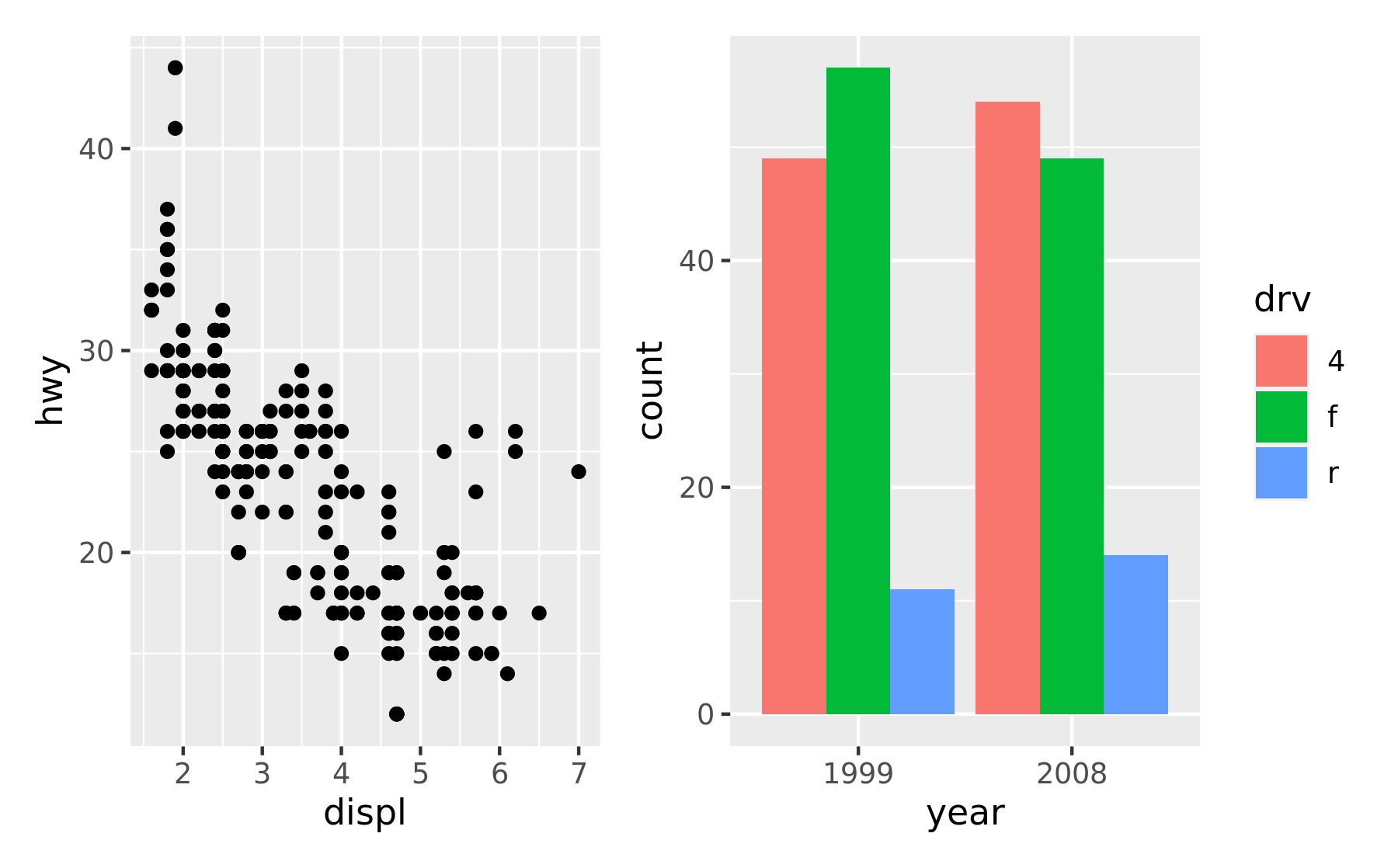
+ does not specify any specific layout, only that the plots should be displayed together. In the absence of a layout the same algorithm that governs the number of rows and columns in facet_wrap() will decide the number of rows and columns. This means that adding 3 plots together will create a 1x3 grid while adding 4 plots together will create a 2x2 grid.
p1 + p2 + p3 + p4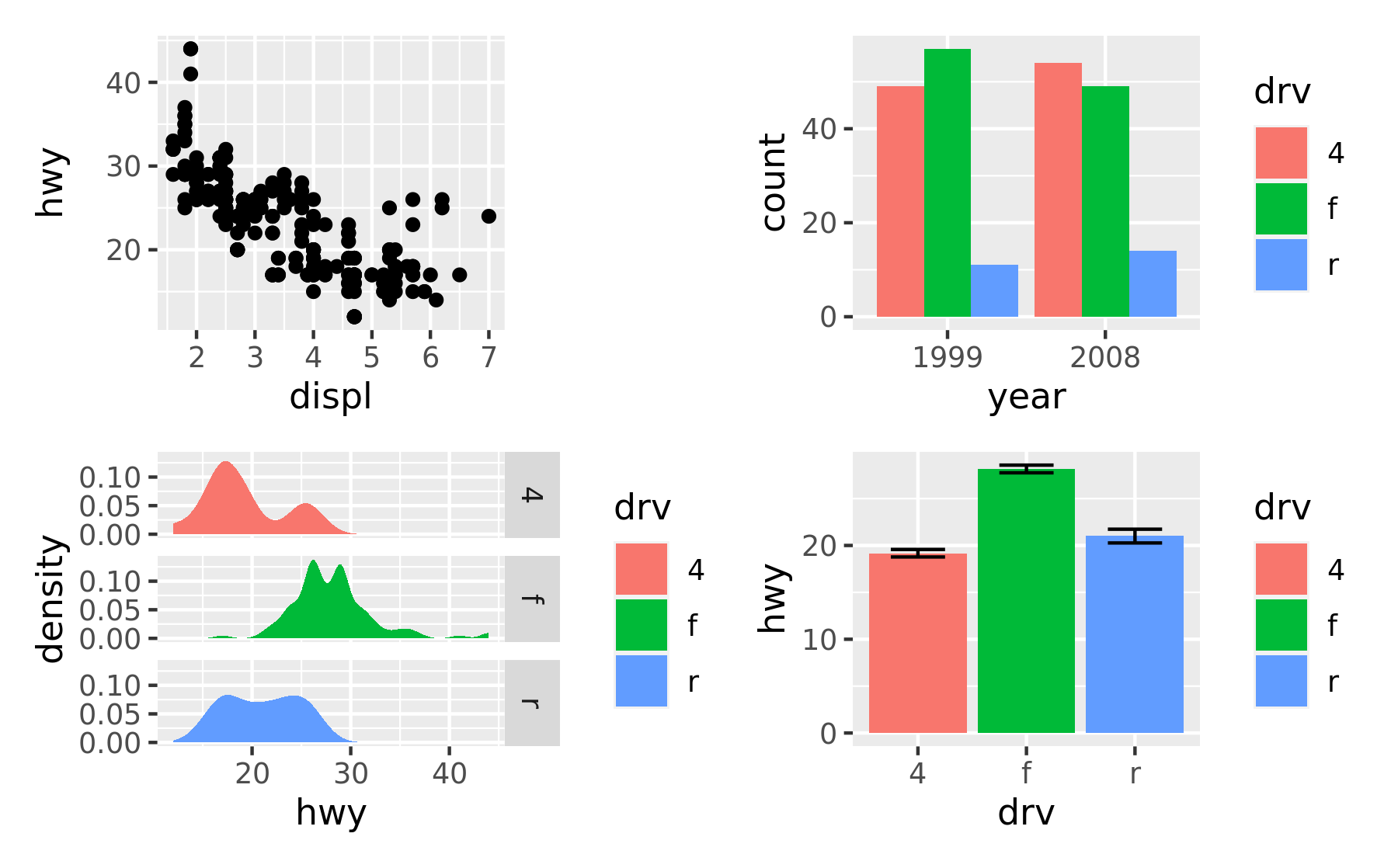
As can be seen from the two examples above, patchwork takes care of aligning the different parts of the plots with each other. You can see that all plotting regions are aligned, even in the presence of faceting. Further, you can see that the y-axis titles in the two left-most plots are aligned despite the axis text in the bottom left plot being wider.
8.1.1 Taking control of the layout
It is often that the automatically created grid is not what you want and it is of course possible to control it. The most direct and powerful way is to do this is to add a plot_layout() specification to the plot:
p1 + p2 + p3 + plot_layout(ncol = 2)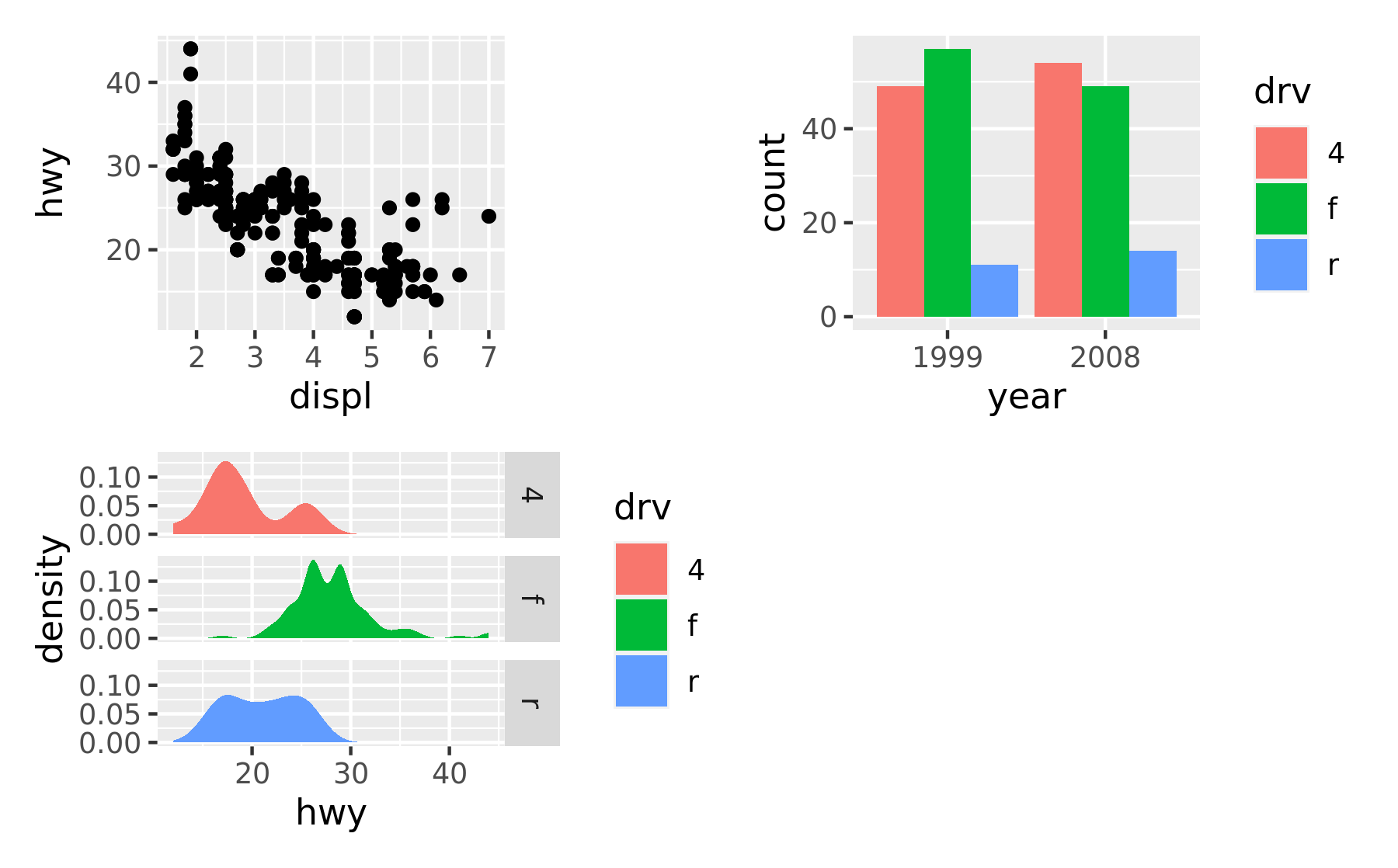
A common scenario is wanting to force a single row or column. patchwork provides two operators, / and | respectively, to facilitate this (under the hood they simply set number of rows or columns in the layout to 1).
p1 / p2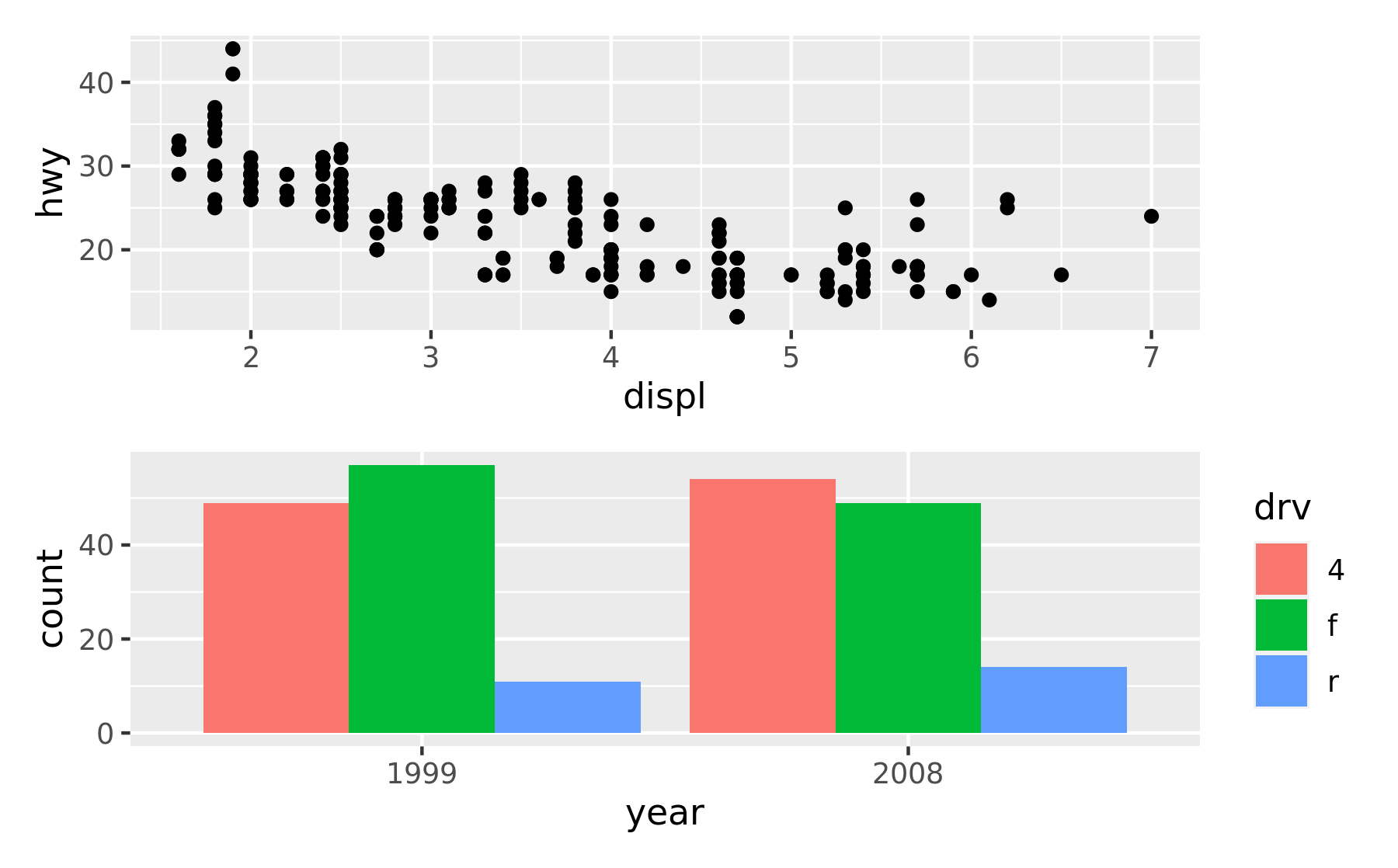
# Basically the same as using `+` but the intend is clearer
p3 | p4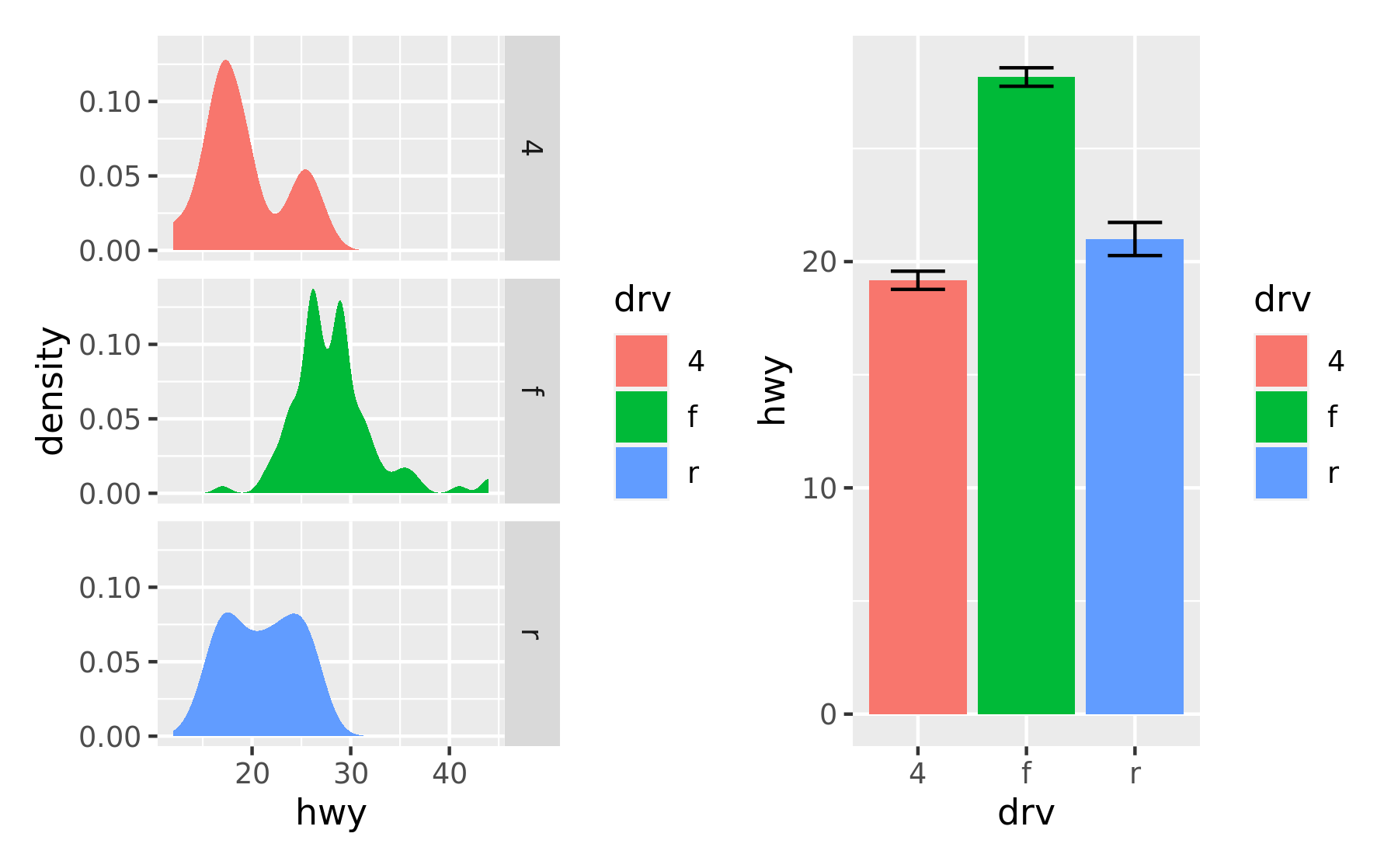
patchwork allows nesting layouts which means that it is possible to create some very intricate layouts using just these two operators
p3 | (p2 / (p1 | p4))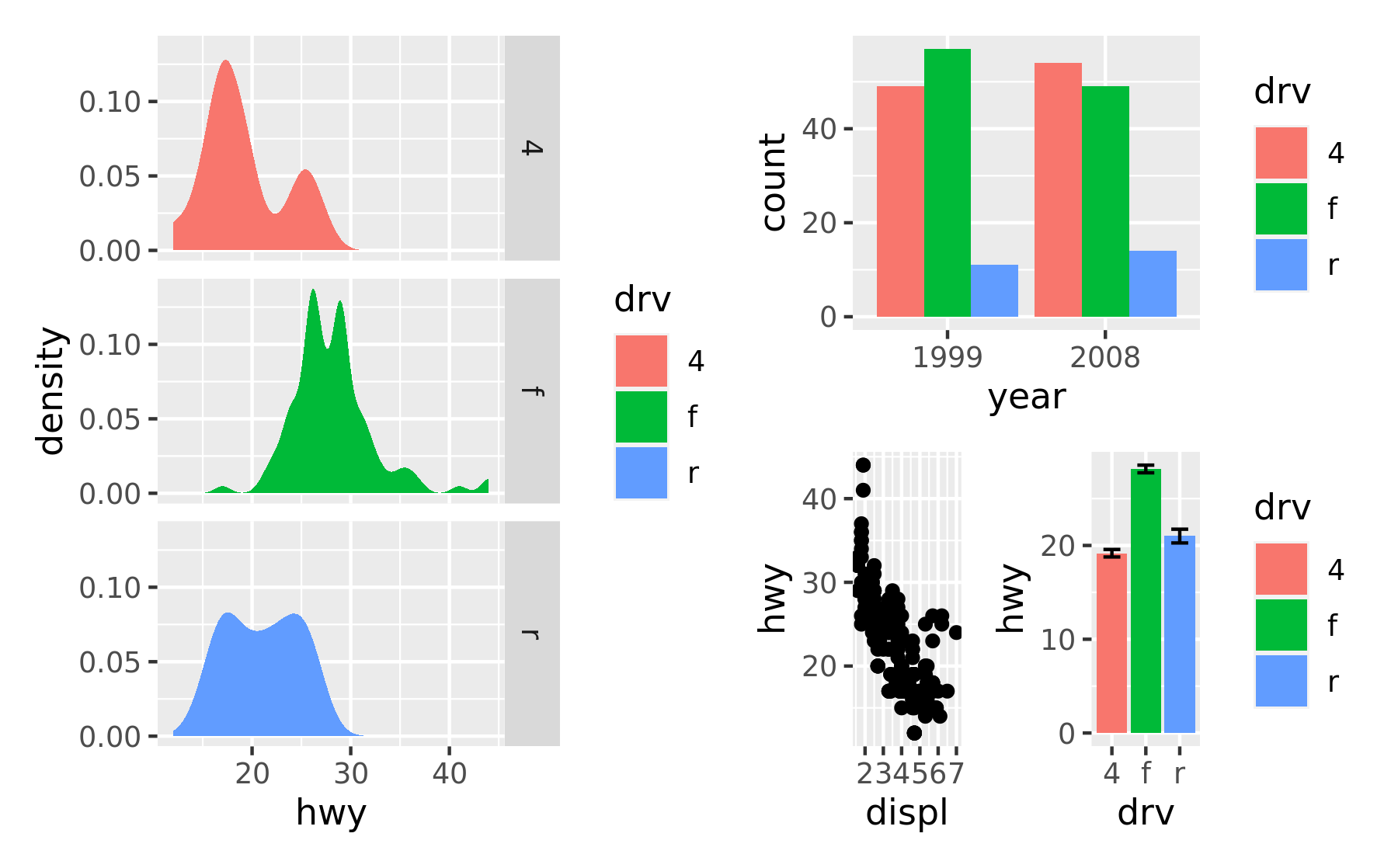
Alternatively, for very complex layouts, it is possible to specify non-tabular layouts with a textual representation in the design argument in plot_layout().
layout <- "
AAB
C#B
CDD
"
p1 + p2 + p3 + p4 + plot_layout(design = layout)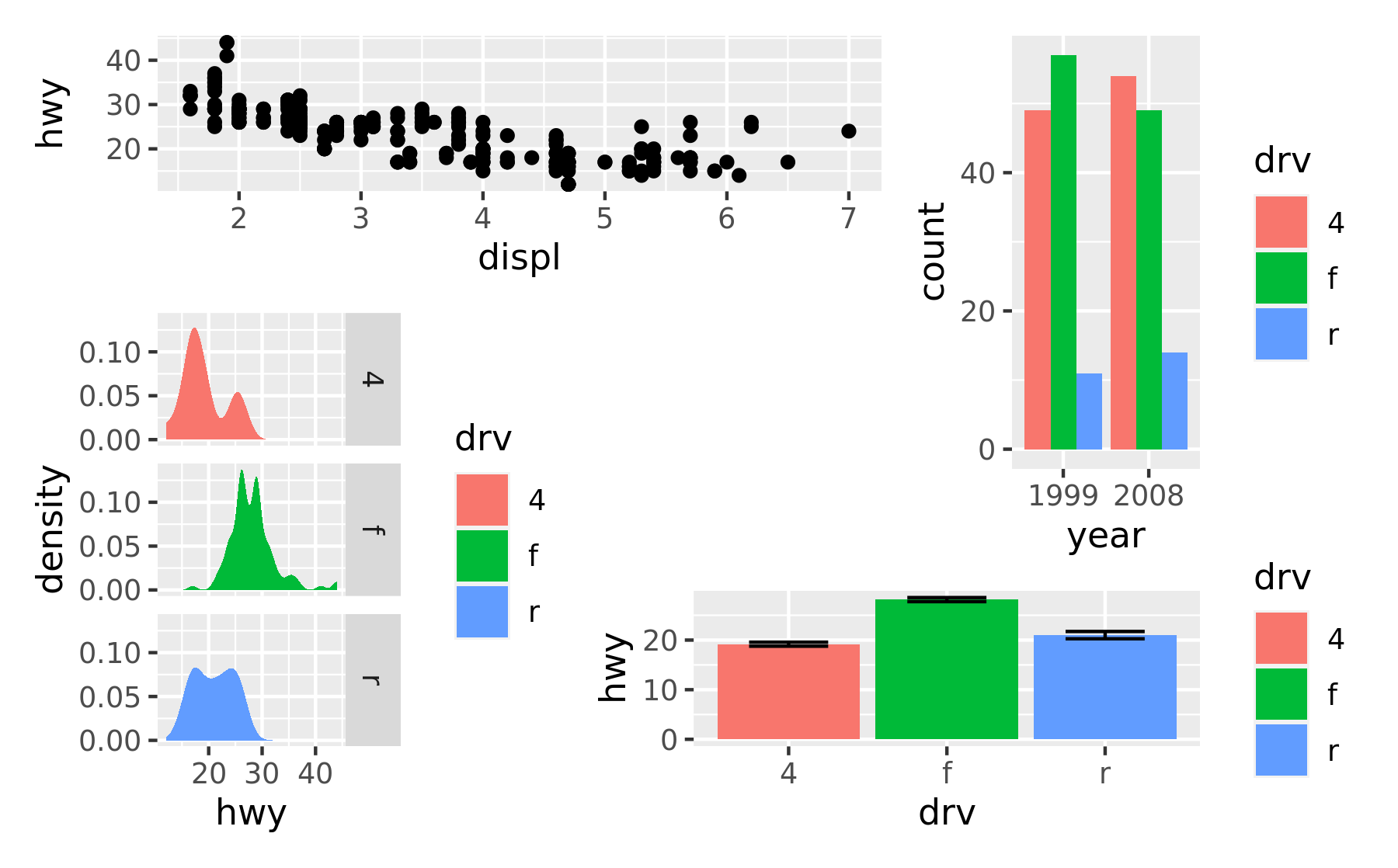
As has been apparent in the last couple of plots the legend often becomes redundant between plots. While it is possible to remove the legend in all but one plot before assembling them, patchwork provides something easier for the common case:
p1 + p2 + p3 + plot_layout(ncol = 2, guides = "collect")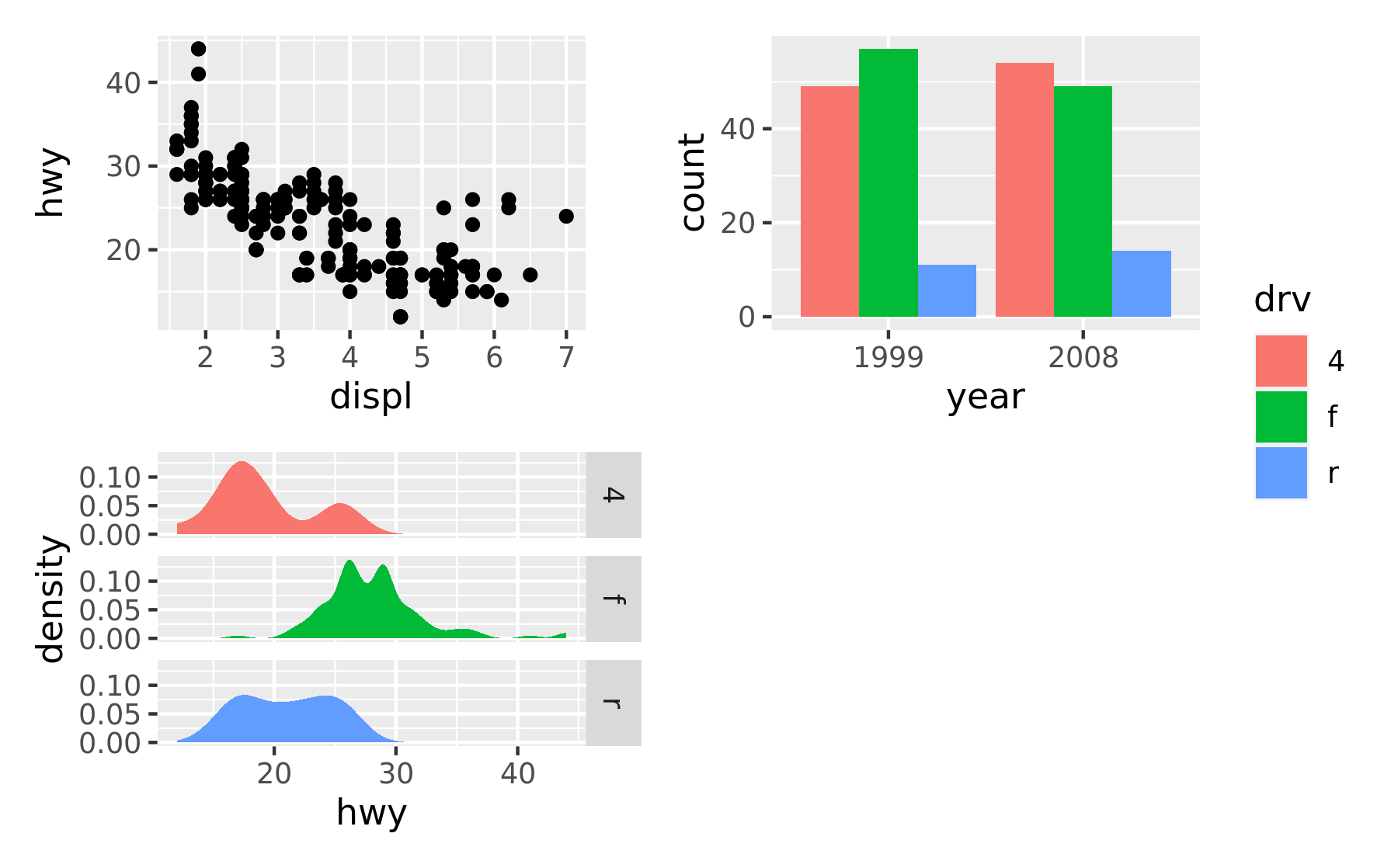
Electing to collect guides will take all guides and put them together at the position governed by the global theme. Further, it will remove any duplicate guide leaving only unique guides in the plot. The duplication detection looks at the appearance of the guide, and not the underlying scale it comes from. Thus, it will only remove guides that are exactly alike. If you want to optimize space use by putting guides in an empty area of the layout, you can specify a plotting area for collected guides:
p1 + p2 + p3 + guide_area() + plot_layout(ncol = 2, guides = "collect")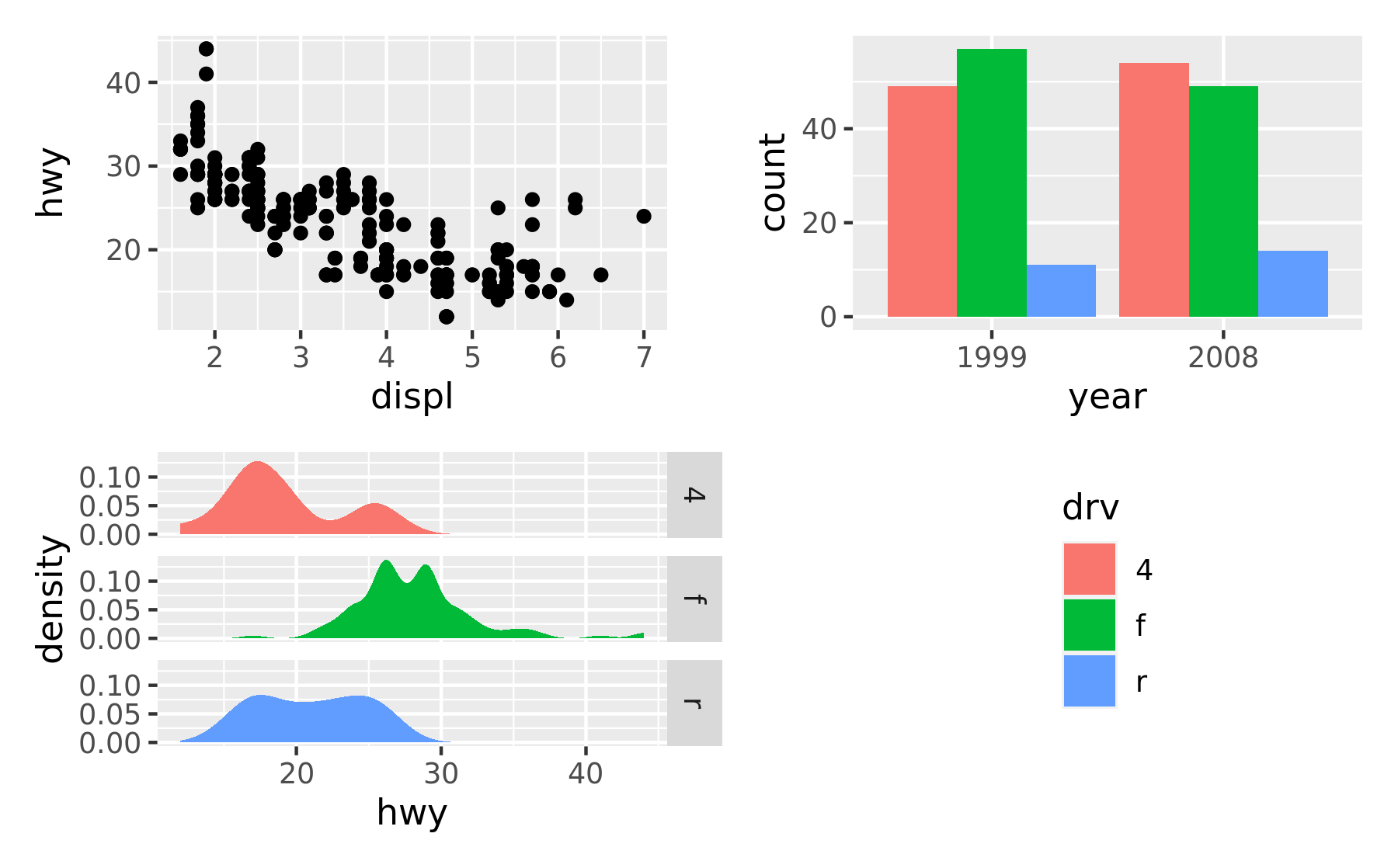
8.1.2 Modifying subplots
One of the tenets of patchwork is that the plots remain as standard ggplot objects until rendered. This means that they are amenable to modification after they have been assembled. The specific plots can by retrieved and set with [[]] indexing:
p12 <- p1 + p2
p12[[2]] <- p12[[2]] + theme_light()
p12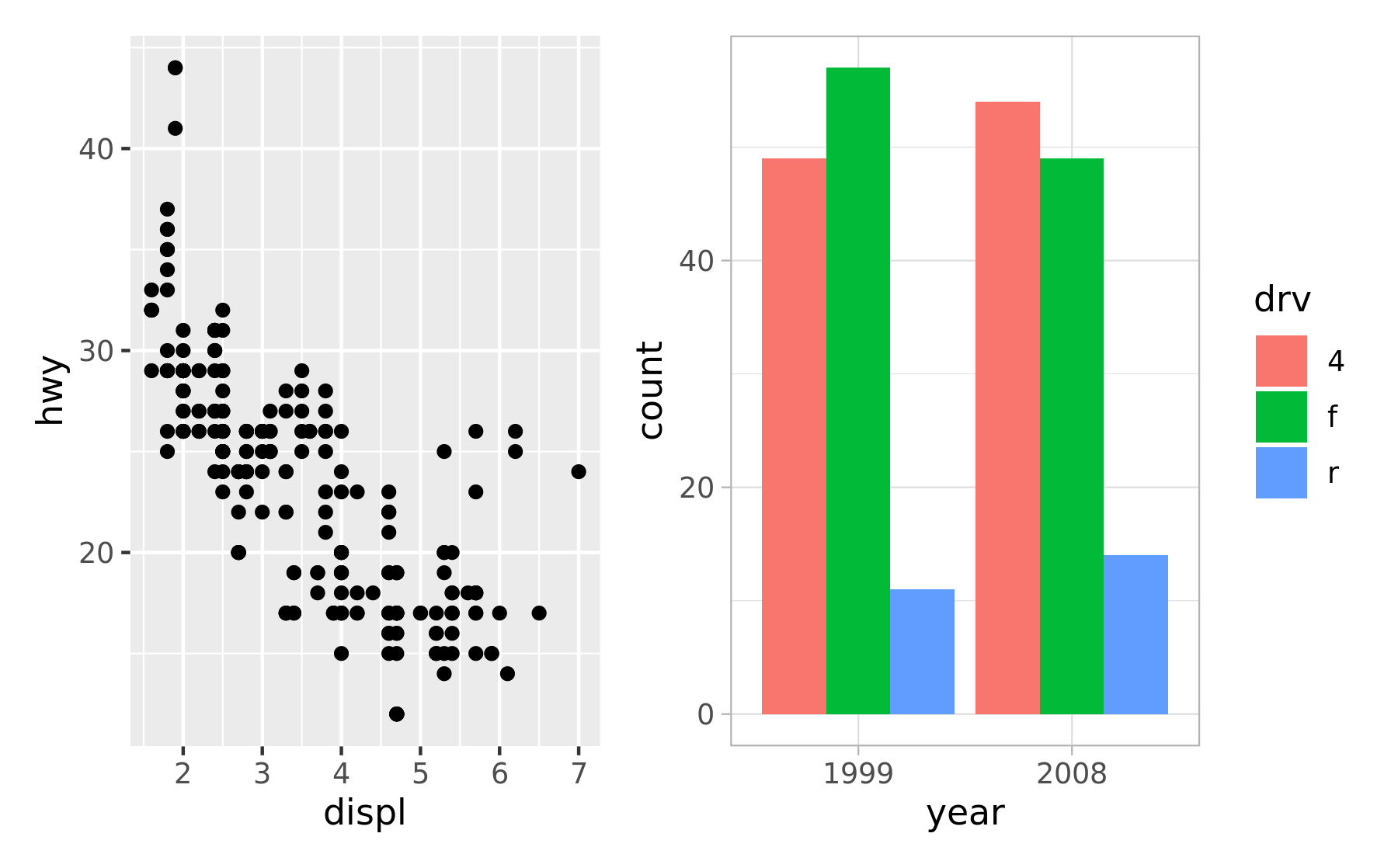
Often though, it is necessary to modify all subplots at once to e.g. give them a common theme. patchwork provides the & for this scenario:
p1 + p4 & theme_minimal()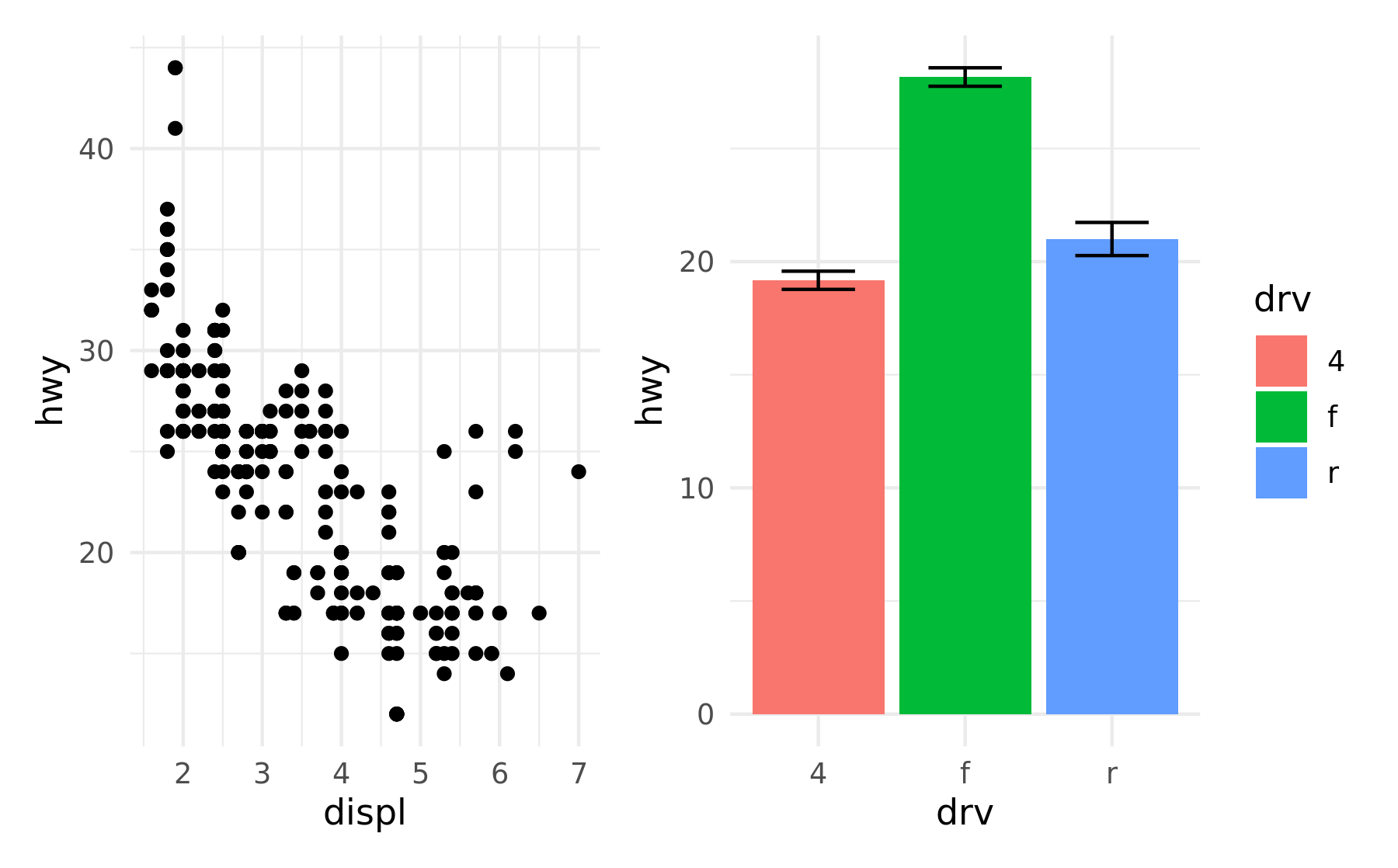
This can also be used to give plots a common axis if they share the same aesthetic on that axis:
p1 + p4 & scale_y_continuous(limits = c(0, 45))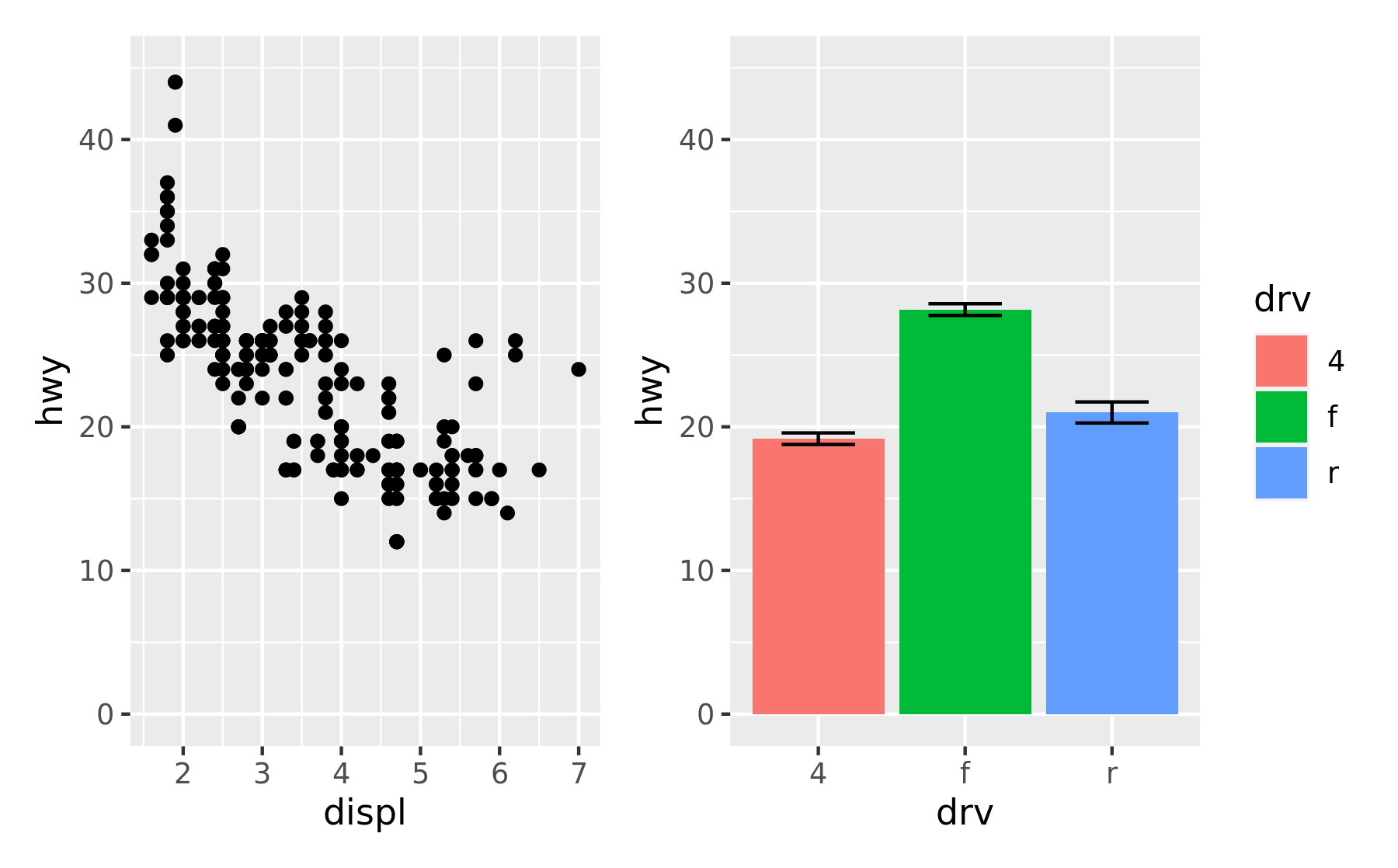
8.1.3 Adding annotation
Once plots have been assembled they start to form a single unit. This also means that titles, subtitles, and captions will often pertain to the full ensemble and not individual plots. Titles etc. can be added to patchwork plots using the plot_annotation() function.
p34 <- p3 + p4 + plot_annotation(
title = "A closer look at the effect of drive train in cars",
caption = "Source: mpg dataset in ggplot2"
)
p34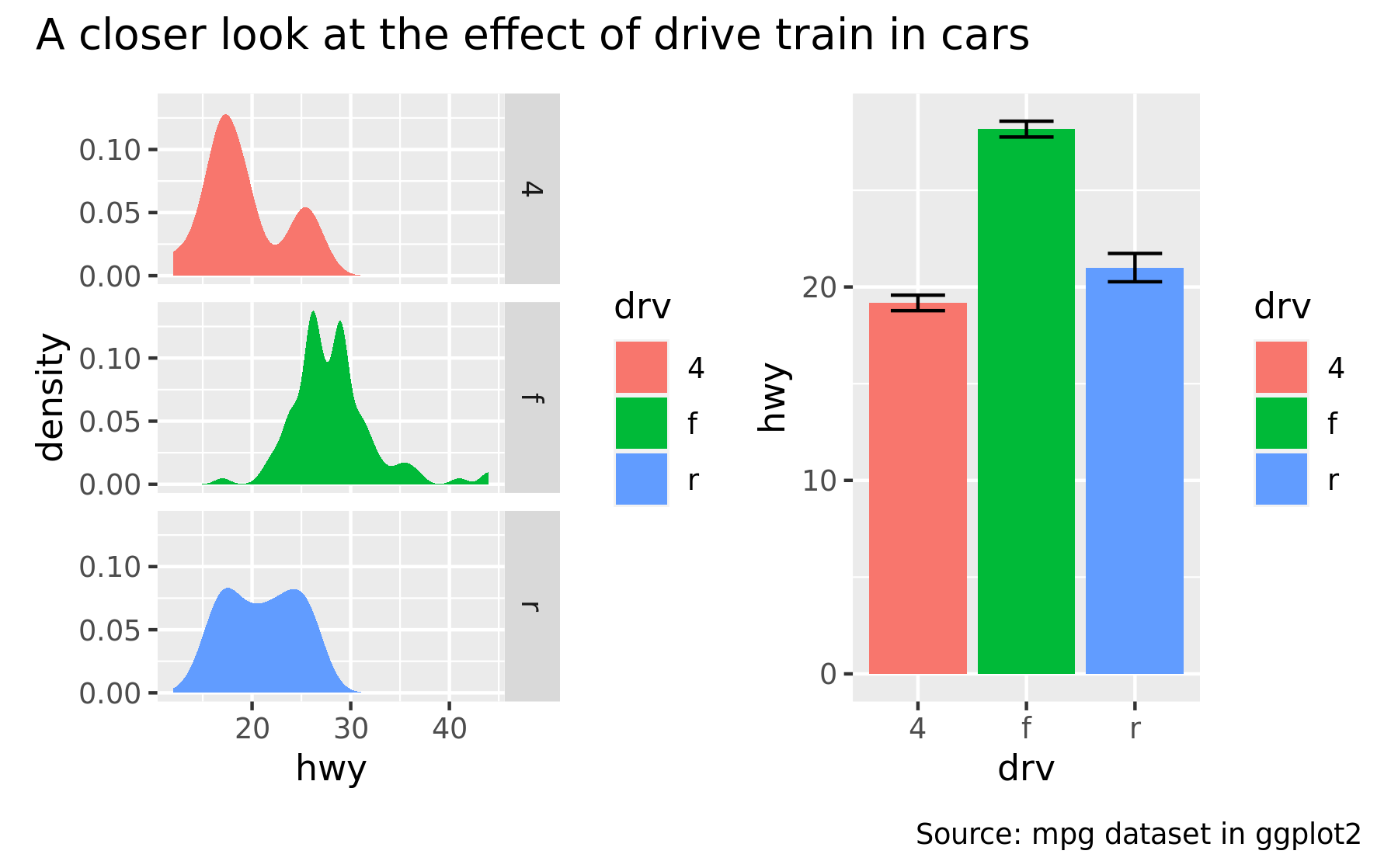
The titles formatted according to the theme specification in the plot_annotation() call.
p34 + plot_annotation(theme = theme_gray(base_family = "mono"))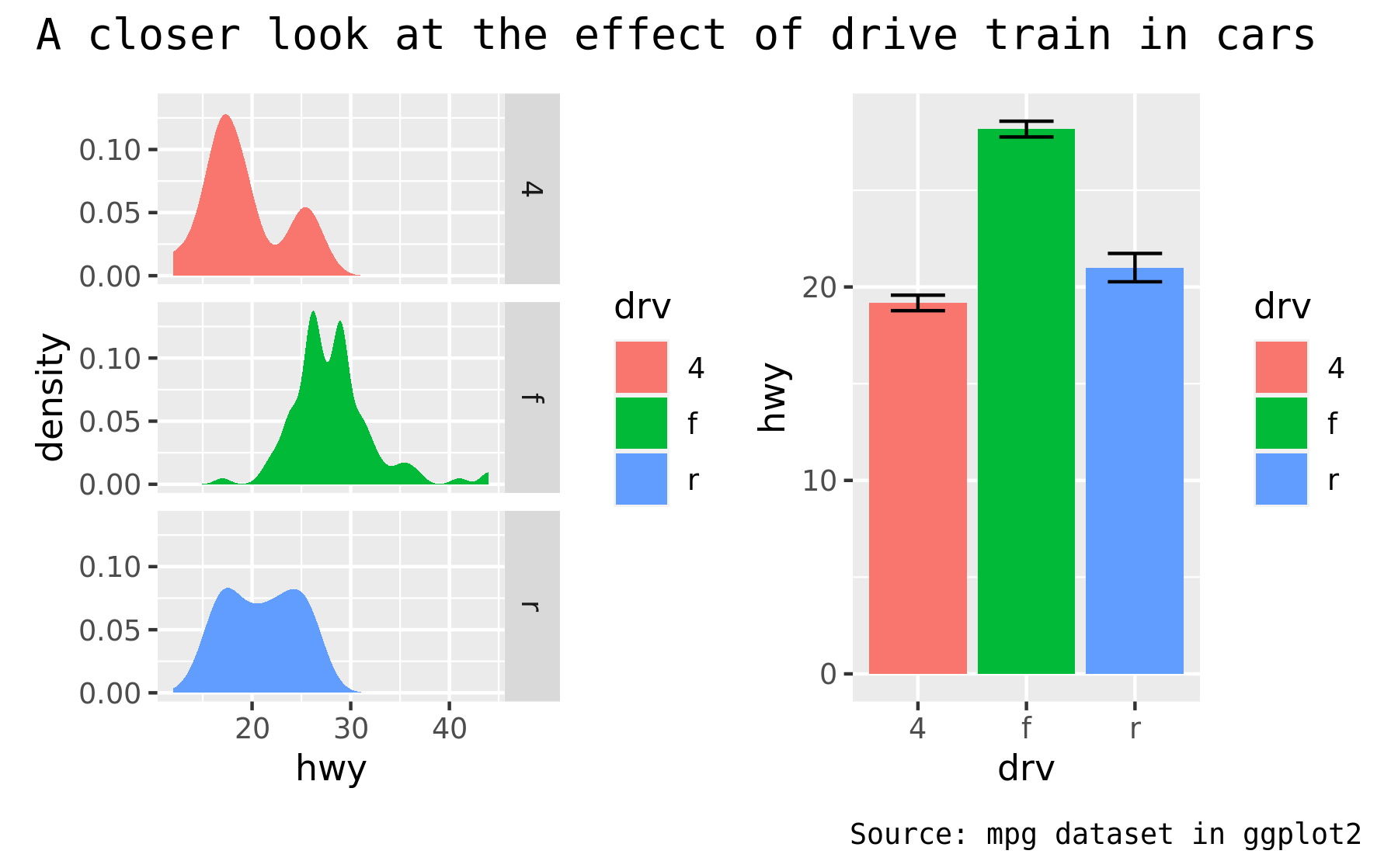
As the global theme often follows the theme of the subplots, using & along with a theme object will modify the global theme as well as the themes of the subplots
p34 & theme_gray(base_family = "mono")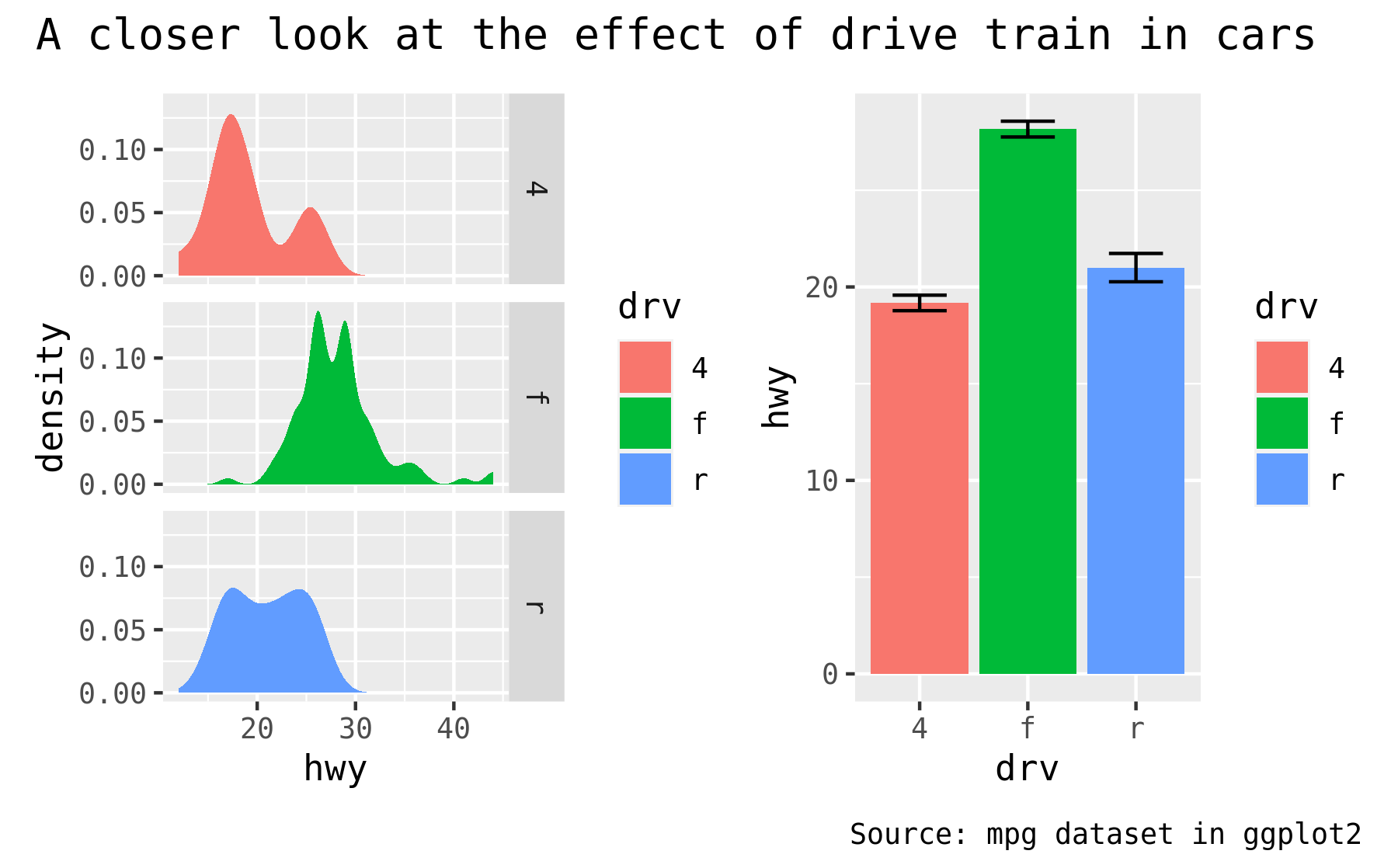
Another type of annotation, known especially in scientific literature, is to add tags to each subplot that will then be used to identify them in the text and caption. ggplot2 has the tag element for exactly this and patchwork offers functionality to set this automatically using the tag_levels argument. It can generate automatic levels in latin characters, arabic numerals, or roman numerals
p123 <- p1 | (p2 / p3)
p123 + plot_annotation(tag_levels = "I") # Uppercase roman numerics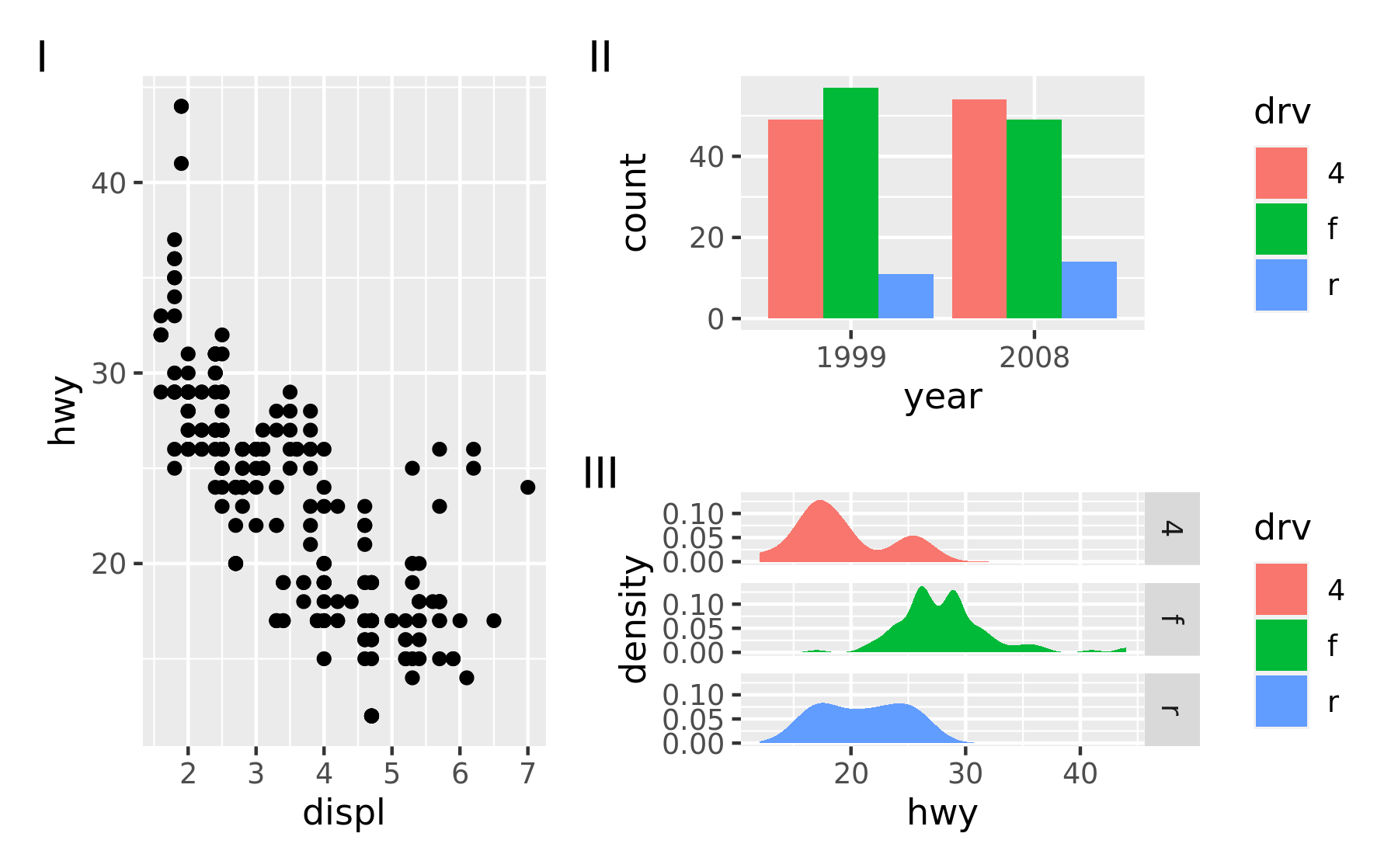
An additional feature is that it is possible to use nesting to define new tagging levels:
p123[[2]] <- p123[[2]] + plot_layout(tag_level = "new")
p123 + plot_annotation(tag_levels = c("I", "a"))
As can be seen, patchwork offers a long range of possibilities when it comes to arranging plots, and the API scales with the level of complexity of the assembly, from simply using + to place multiple plots in the same area, to using nesting, layouts, and annotations to create advanced custom layouts.